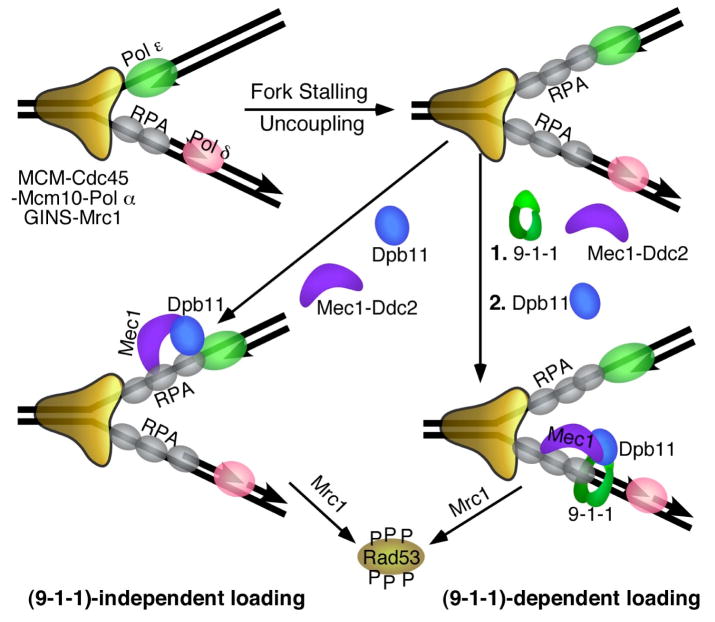
Figure 4. The DNA replication checkpoint.
S. cerevisiae proteins that are involved in checkpoint function are shown. See Table 1 for the homologous human proteins. Stalling of the fork results in uncoupling of the leading and lagging strands, and in the formation of extended RPA-coated ssDNA regions that recruit Mec1. The 9-1-1 clamp is shown loaded onto the 5′-junction of an Okazaki fragment because of its demonstrated loading specificity, but might also load onto the leading strand after fork restart by Pol α-primase [90]. Recruitment of Dpb11/TopBP1/Cut5 to the fork is facilitated through interactions with 9-1-1 ((9-1-1)-dependent loading, right). In S. cerevisiae, (9-1-1)-independent recruitment of Dpb11 may also occur, possibly through interactions with Pol ε. Dpb11/TopBP1/Cut5 activates Mec1/ATR to phosphorylate Rad53/Chk1 and additional factors. Mrc1/Claspin is the proposed downstream chromatin-associated mediator that activates transphosphorylation of Rad53/Chk1 [91].