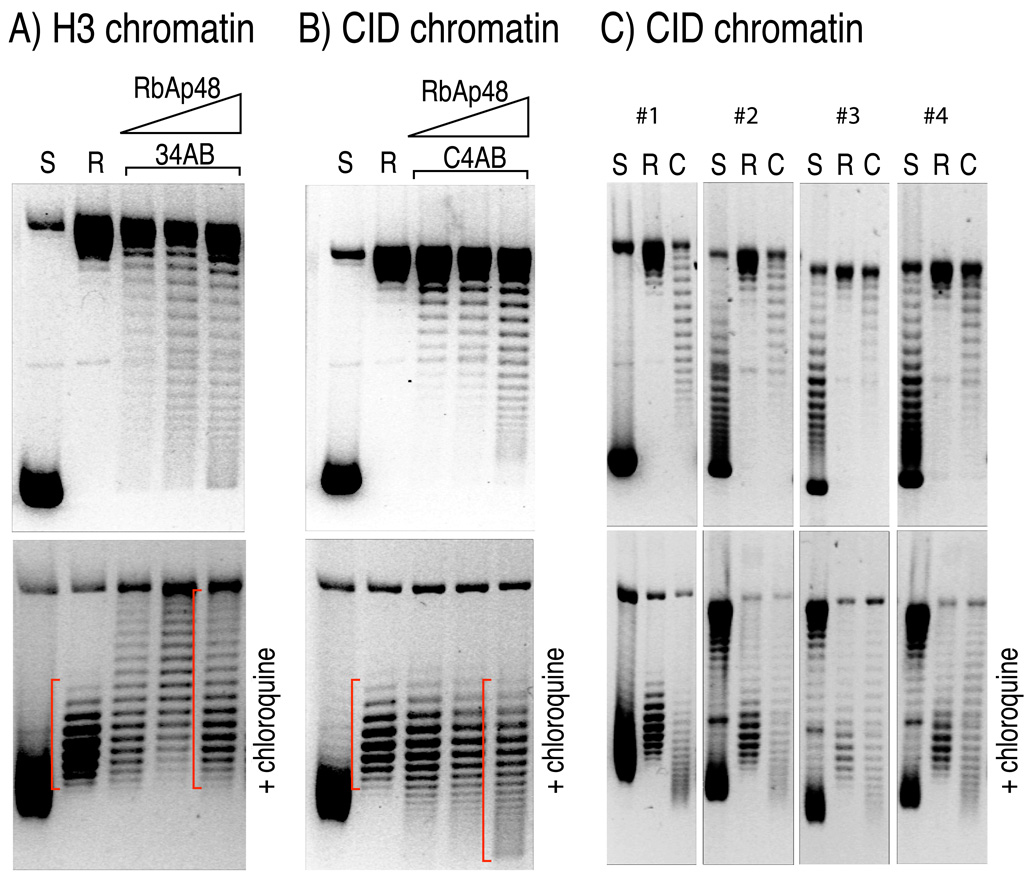
Figure 1. Drosophila CID chromatin assembled in vitro induces positive supercoils in closed circular plasmids.
(A) An ~7 kb plasmid containing an ~3 kb Drosophila satellite DNA insert (Furuyama et al., 2006) electrophoreses as negatively supercoiled species in an agarose gel after isolation from E.coli (S). The plasmid DNA was relaxed by topoisomerase (R), and H3 chromatin was assembled in vitro (34AB) in the presence of the Drosophila RbAp48 chaperone, as previously described. The top panel shows the topoisomer separation on an agarose gel without chloroquine, whereas the bottom panel shows their migration in the presence of 1µg/ml chloroquine. Gels were stained with ethidium bromide after separation to visualize DNA. The slower migration of topoisomers induced by H3-containing nucleosome relative to the migration of relaxed DNA (R) indicates that H3-nucleosomes induce negative supercoils (red bracket). (B) same as in (A), except that CID (Drosophila CenH3) was used in place of H3 (C4AB). The faster migration of topoisomers induced by CID-containing nucleosome relative to the migration of relaxed DNA (R) in the presence of chloroquine indicates that CID-nucleosomes induce positive supercoils in vitro. (C) Identical chromatin assembly reactions were performed as in (B) using four independent plasmids containing randomly cloned 3-kb Drosophila genomic DNA (Clone #1 through #4; See also Figure S1). CID nucleosomes induce positive supercoils regardless of DNA sequence, as evident from migration patterns that are similar to those in (B). S: negatively supercoiled; R, relaxed; C: CID chromatin.