Fig. 2.
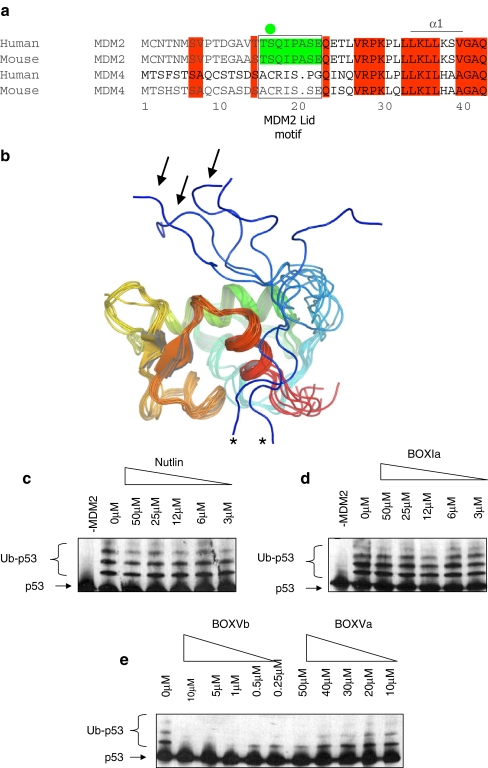
The N-terminal pseudo-substrate motif of MDM2. a Conservation of amino acids 1–44, including the pseudo-substrate motif (aka, the lid) within MDM2 homologues and absence of the pseudo-substrate motif in the orthologue MDM4. The pseudo-substrate motif is surrounded by a box, and the phosphorylation site at Ser17 is highlighted with a green circle. b The NMR structure of the non-liganded N-terminal p53 binding domain of MDM2 (PDB ID 1Z1M, residues 10–109; apo-MDM2 [45]) reveals a large degree of conformational heterogeneity in the N-terminal 20 residues (colored from blue (N-terminal) to red (C-terminal)). Pseudo-substrate motif conformations acting like a lid to cover the MDM2 hydrophobic pocket are highlighted with arrows, and conformations with the pseudo-substrate motif interacting with the MDM2 surface are highlighted with asterisks. c–e The effects of small ligands on MDM2 function as an E3 ubiquitin ligase. A ubiquitination assay was assembled with purified E1, E2, MDM2, and p53 proteins without or with increasing concentrations of c Nutlin, dBOX-I peptide derived from the N-terminus of p53, or eBOX-V homology peptides derived from Rb (BOX-Vb) and p53 DNA binding domain (BOX-Va). Ubiquitination was assayed as indicated in the “Materials and methods”. The position of the un-ubiquitinated p53 substrate is highlighted with arrows and p53-ubiquitin adducts with brackets