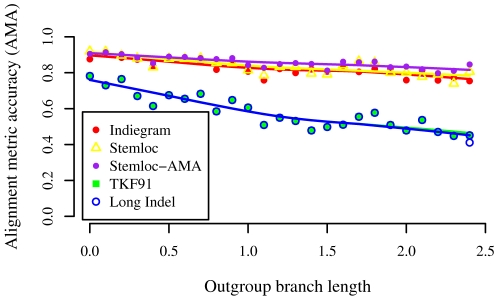
Figure 11. Dependence of alignment metric accuracy on alignment method and outgroup branch length.
We simulated the evolution of three structural RNAs under the TKFST model. The simulation included two sister species at unit distance from the ancestral sequence, plus one “outgroup” whose branch length  was varied between
was varied between 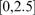 by selecting 25 equally spaced values of
by selecting 25 equally spaced values of  in this range, spaced
in this range, spaced 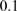 apart. We then simulated 25 alignments for each value of
apart. We then simulated 25 alignments for each value of  , using TKFST model parameters described in the text. The Alignment Metric Accuracy (AMA) is, roughly, the proportion of residues that are correctly aligned, averaged over all pairs of sequences (see [54] for a precise definition; we set the AMA Gap Factor to 1). The AMA between the true alignment and the inferred alignment was measured for various statistical alignment inference procedures. These procedures are described in the text, but may be summarized very briefly as ML under the true model (“Indiegram”); greedy approximate-ML progressive alignment by single-linkage clustering with pair SCFGs (“Stemloc”); sequence annealing, a form of posterior decoding to maximize a sum-over-pairs accuracy metric, using pair SCFGs to get the posterior probabilities (“Stemloc-AMA”); statistical alignment using the TKF91 model, i.e. linear gap-penalties (“TKF91”); and statistical alignment using a long-indel model, i.e. affine gap-penalties (“Long Indel”).
, using TKFST model parameters described in the text. The Alignment Metric Accuracy (AMA) is, roughly, the proportion of residues that are correctly aligned, averaged over all pairs of sequences (see [54] for a precise definition; we set the AMA Gap Factor to 1). The AMA between the true alignment and the inferred alignment was measured for various statistical alignment inference procedures. These procedures are described in the text, but may be summarized very briefly as ML under the true model (“Indiegram”); greedy approximate-ML progressive alignment by single-linkage clustering with pair SCFGs (“Stemloc”); sequence annealing, a form of posterior decoding to maximize a sum-over-pairs accuracy metric, using pair SCFGs to get the posterior probabilities (“Stemloc-AMA”); statistical alignment using the TKF91 model, i.e. linear gap-penalties (“TKF91”); and statistical alignment using a long-indel model, i.e. affine gap-penalties (“Long Indel”).