Abstract
A lysine racemase (lyr) gene was isolated from a soil metagenome by functional complementation for the first time by using Escherichia coli BCRC 51734 cells as the host and d-lysine as the selection agent. The lyr gene consisted of a 1,182-bp nucleotide sequence encoding a protein of 393 amino acids with a molecular mass of about 42.7 kDa. The enzyme exhibited higher specific activity toward lysine in the l-lysine-to-d-lysine direction than in the reverse reaction.
Amino acids are the building blocks of proteins and play an important role in the regulation of the metabolism of living organisms. Among two enantiomers of naturally occurring amino acids, l-amino acids are predominant in living organisms, while d-amino acids are found in both free and bound states in various organisms like bacteria (36), yeasts (35), plants (47), insects (11), mammals (17), bivalves (39), and fish (28). The d-amino acids are mostly endogenous and produced by racemization from their counterparts by the action of a racemase. Thus, the amino acid racemases are involved in d-amino acid metabolism (29, 46). Since the discovery of alanine racemase in 1951 (42), several racemases toward amino acids, such as those for glutamate, threonine, serine, aspartate, methionine, proline, arginine, and phenylalanine, have been reported in bacteria, archaea, and eukaryotes, including mammals (1, 2, 15, 30, 31, 44). They are classified into two groups: pyridoxal 5′-phosphate (PLP)-dependent and PLP-independent enzymes (9, 36).
Lysine racemase (Lyr, EC 5.1.1.5) was first reported in Proteus vulgaris ATCC 4669 (19) and proposed to be involved in the lysine degradation of bacterial cells (5, 19). Catabolism of lysine occurs via two parallel pathways. In one of the pathways, δ-aminovalerate is the key metabolite, whereas in the other l-lysine is racemized to d-lysine, and l-pipecolate and α-aminoadipate (AMA) are the key metabolites (5). d-Lysine catabolism proceeds through a series of cyclized intermediates which are necessary to regenerate an α-amino acid and comprise the following metabolites (AMA pathway): d-lysine→α-keto-ɛ-amino caproate→Δ1-piperideine-2-carboxylate→pipecolate→Δ1-piperideine-6-carboxylate→α-amino-δ-formylcaproate→α-AMA→α-ketoadipate (6, 7, 12, 27). The final product is converted to α-ketoglutarate via a series of coenzyme A derivatives and subsequently participates as an intermediate in the Krebs cycle. This pathway suggests that the biological function of d-lysine in the bacteria is that of a carbon or nitrogen source. Racemization of added l-lysine to d-lysine by whole cells of Proteus spp. and Escherichia spp. (19) and by the cell extract of Pseudomonas putida ATCC 15070 (5, 20) has been found. However, the enzyme has not been purified to homogeneity, and thus, its molecular and catalytic characteristics, including its gene structure, have not been elucidated. In this study, we explored a metagenomic library constructed from a garden soil to isolate a novel Lyr enzyme. After expression in Escherichia coli, the purified enzyme was characterized in terms of optimal pH and temperature, thermal stability, and racemization activity.
Isolation of lysine racemase gene from a soil metagenome.
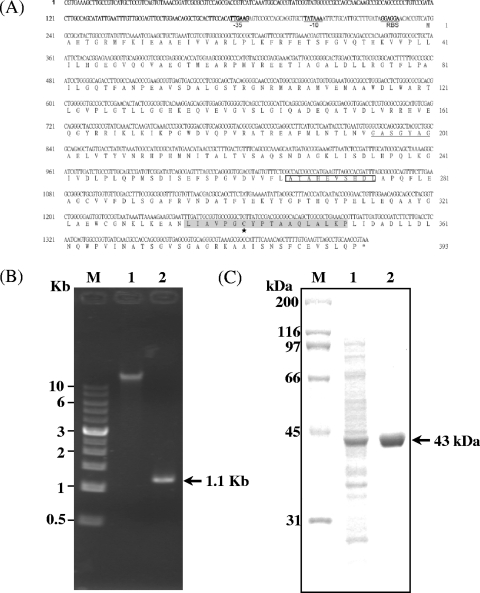
We constructed a soil metagenomic library in plasmid pUC18 by using the genomic DNA isolated from the garden soil of the National Chung Hsing University (Taichung, Taiwan). Recombinant plasmids were introduced into lysine-auxotrophic mutant E. coli BCRC 51734 and screened for the transformants on an M9 plate supplemented with d-lysine. One E. coli transformant harboring the recombinant plasmid, designated pUC-m19lyr, was obtained. DNA sequence analysis of the insert DNA identified an open reading frame encoding a protein composed of 393 amino acids with a calculated molecular mass of approximately 42.7 kDa. A putative ribosomal binding site (GGAGGA) was found to be located 8 bp upstream of the ATG start codon, and potential −35 (TTGAAG) and −10 (TATAAA) consensus promoter sequences were also observed (Fig. 1A). In order to confirm that lyr did not evolve from the ligation of Sau3AI-digested DNA fragments during the preparation of the metagenomic library in E. coli cells and actually exists in the metagenome from the original soil sample, we designed primers based on the gene sequence to amplify the lyr gene by the PCR method. The amplified product, with an expected size of 1.1 kb and corresponding to the cloned lyr gene, suggested the presence of lyr in the metagenomic DNA (Fig. 1B).
FIG. 1.
Identification and expression of a novel lysine racemase gene from a soil metagenomic library. (A) Nucleotide and amino acid sequences of the lyr gene. The putative promoter regions (−35 and −10 regions) and the ribosomal binding site (RBS) are in boldface and underlined. The asterisk denotes the stop codon. The residues of a putative NADP-binding motif (GxxGxxG, underlined), a zinc-binding region (open box), a catalytic region (gray box), and a catalytic site (black star) are shown. (B) PCR amplification of the lyr gene from soil metagenomic DNA. Lane M, size marker; lane 1, soil metagenomic DNA; lane 2, lyr gene amplified from soil metagenomic DNA. (C) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of Lyr expressed by IPTG-induced E. coli JM109(pQE-lyr). Lane M, protein standards; lane 1, crude extract of IPTG-induced cells; lane 2, purified Lyr.
Biochemical properties of Lyr.
To express His6-tagged Lyr in E. coli, the lyr gene from a Lyr-producing transformant was amplified by PCR and subcloned into pQE30 between the BamHI and HindIII sites (Qiagen, Valencia, CA) and expressed under the control of the lacZ promoter. Expression of the gene was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubation at 28°C for 6 h. The recombinant Lyr protein appeared as a single major band with an expression level of approximately 20% of the total cell proteins. The purified Lyr protein was quantified with the commercial Bradford protein assay kit (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as the standard. About 2 mg of Lyr could be purified from 500 ml of culture broth by nickel chelate affinity chromatography (Qiagen). Purified Lyr exhibited an apparent molecular mass of about 43 kDa as estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, which was in close agreement with the calculated mass of 42.7 kDa of the Lyr protein (Fig. 1C).
Lyr activity was determined by measuring the production of d-lysine from l-lysine or vice versa by enantioselective column chromatography. The reaction mixture (1 ml), containing an appropriate amount of purified enzyme, 50 mM Tris-HCl buffer (pH 8.0), 2 mM cobalt chloride, and 10 mM d- or l-lysine, was incubated at 30°C for 30 min, and the reaction was terminated by heating at 95°C for 5 min. The production of d- or l-lysine was analyzed by high-performance liquid chromatography on a 150- by 4.0-mm Crownpak CR(+) column (Daicel Chemical Industries Ltd., Tokyo, Japan) at a flow rate of 0.4 ml/min. The mobile phase used was 5% methanol and 95% aqueous HClO4 (pH 1.5), and the detection wavelength was set at 200 nm. One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmol of l-lysine or d-lysine per min under the assay conditions used. We used a Crownpak CR(+) chiral column for high-performance liquid chromatography because it had been successfully used previously in the resolution of amino acids (25, 43). Using this method, we were able to clearly separate and detect d- and l-lysine with high sensitivity (see Fig. S1 and S2 in the supplemental material). Lyr exhibited higher specific activity (3.61 versus 1.68 U/mg protein), as well as a higher kcat (0.085 ± 0.003 versus 0.036 ± 0.002 min−1), in the l-lysine-to-d-lysine direction than in the reverse reaction. The apparent Km values of purified Lyr were determined to be 16.21 ± 0.26 and 23.48 ± 1.22 mM for l- to d-lysine and d- to l-lysine, respectively, at 30°C and l-lysine or d-lysine concentrations ranging from 0.1 to 20 mM in Tris-HCl buffer (pH 8.0).
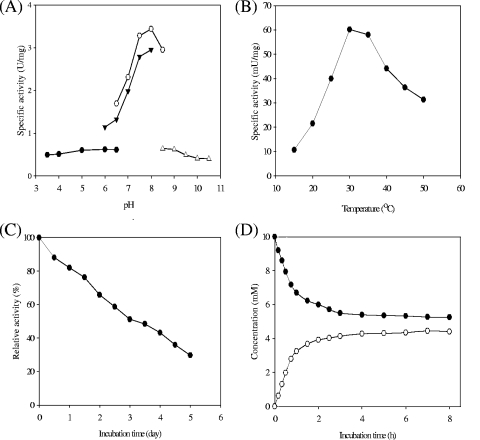
The optimal pH was observed to be around 8.0 in Tris-HCl buffer when the enzyme activity was determined at various pHs (3.5 to 10.5) with different buffers containing 10 mM l-lysine, and only about 15% of maximal activity was observed at pHs of ≤5 and ≥9 (Fig. 2A). The activity of the enzyme was maximum at 30°C when assayed under standard conditions at temperatures ranging from 15 to 50°C (Fig. 2B). The enzyme showed considerable stability at 50°C, with a half-life of about 3 days (Fig. 2C).
FIG. 2.
Biochemical properties of Lyr. (A) Effect of pH on the racemization activity of Lyr toward l-lysine. The buffers used were 50 mM citric acid-Na2HPO4, pH 3.5 to 6.5 (•); 50 mM Tris-HCl, pH 6.5 to 8.5 (○); 50 mM K2HPO4-KH2PO4, pH 6.0 to 8.0 (▴); and 50 mM glycine-NaOH, pH 8.5 to 10.5 (▵). (B) Effect of temperature on racemization activity. (C) Thermostability of purified Lyr. Enzyme activities were assayed after the enzyme was incubated at 50°C. (D) Conversion of l-lysine to d-lysine by recombinant Lyr. Symbols: •, l-lysine; ○, d-lysine.
We analyzed the bound metal ions in purified Lyr with an inductively coupled plasma spectrometer (GBC Scientific Equipment, Australia). The dialyzed enzyme (2 mg/ml) was used for the analysis, and the amount of ion was calculated by using the standard solution for each ion. The results revealed that dialyzed Lyr contained 0.77 mol of Zn2+ per mol of the dimeric enzyme, while cobalt, magnesium, manganese, nickel, or copper ions were absent (data not shown). To determine the effect of metal ions on Lyr activity, about 10 U of purified enzyme was incubated with the chloride salt of metal ions in 50 mM Tris-HCl buffer (pH 8.0) at 30°C for 30 min and demetalized by dialysis against a buffer containing 50 mM Tris-HCl (pH 8.0) with 10 mM EDTA or dipicolinic acid and then against 50 mM Tris-HCl buffer (pH 8.0). The racemization activity of purified Lyr was completely inhibited by metal-chelating agents such as EDTA and dipicolinic acid and could be recovered by the addition of Co2+, Mg2+, Mn2+, or Zn2+ ions (Table 1), indicating that Lyr is a metalloenzyme. The divalent metal ions at a 2 mM concentration obviously affected Lyr activity. The additional extrinsic Co2+, Mg2+, and Mn2+ ions enhanced the enzyme activity up to 114.5, 83.6, and 97.3%, respectively. The enzyme was not affected by the addition of 10 mM PLP or 2 mM hydroxylamine and therefore was distinct from PLP-dependent amino acid racemases. The absorption spectrum of Lyr showed no peaks in the near-UV or visible region, except for the absorption due to tryptophan and tyrosine residues of the protein, showing that the enzyme did not contain PLP. Thus, Lyr is a PLP-independent enzyme. Several amino acid racemases, including glutamate racemase and proline racemase, do not require coenzymes for their activity (4, 29), in which thiol groups of cysteine residues serve as catalytic bases in the proton transfer reactions. However, the alanine racemase could catalyze the racemization of amino acids mediated by PLP as its coenzyme (35, 45, 47). The racemization activity of serine racemases from Schizosaccharomyces pombe and mice can be enhanced by divalent cations such as Ca2+, Mn2+, and Mg2+ in coordination with Glu208 and Asp214 (10). We next determined the conversion ratio of l-lysine to d-lysine by Lyr. In a reaction mixture (500 μl) containing 10 U purified Lyr and 10 mM l-lysine, about 42.8% of the l-lysine was converted to d-lysine in 6 h, and equilibrium of these two isomers could be obtained in a 16-h reaction (Fig. 2D). The spontaneous racemization of l-lysine was very low. When Lyr was not included in the reaction mixture, less than 1% of the l-lysine was converted to d-lysine.
TABLE 1.
Effects of metal ions and chemical compounds on Lyr activity
| Condition | Concn (mM) | Sp act (U mg−1) | Relative activity (%)a |
|---|---|---|---|
| None | 3.61 | 100 | |
| PLP | 10 | 3.56 | 98.7 |
| Hydroxylamine | 2 | 3.58 | 99.2 |
| Hydroxylamine + PLP | 2, 10 | 3.54 | 98.1 |
| EDTA | 10 | 0 | 0 |
| De-EDTAb | 0 | 0.08 | 2.3 |
| De-EDTA + Co2+ | 0, 2 | 7.74 | 214.5 |
| De-EDTA + Mg2+ | 0, 2 | 6.63 | 183.6 |
| De-EDTA + Mn2+ | 0, 2 | 7.11 | 197.3 |
| De-EDTA + Zn2+ | 0, 2 | 4.38 | 121.2 |
| De-EDTA + Ni2+ | 0, 2 | 0.87 | 24.2 |
| De-EDTA + PLP | 0, 10 | 0.20 | 5.5 |
| Dipicolinic acidc | 10 | 0 | 0 |
| De-dipicolinc acid | 0 | 0.16 | 4.4 |
| De-dipicolinic acid + Co2+ | 0, 2 | 6.80 | 188.4 |
| De-dipicolinic acid + Mg2+ | 0, 2 | 4.52 | 125.3 |
Lysine racemase activity assayed without metal ion addition was taken as 100%.
De-EDTA, removal of EDTA by sample dialysis against 50 mM Tris-HCl buffer (pH 8.0).
De-dipicolinic acid, removal of dipicolinic acid by sample dialysis against 50 mM Tris-HCl buffer (pH 8.0).
NAGPR and NAAAR activities of Lyr.
Why does the Lyr protein possess racemization activity toward lysine? Bioinformatic analysis revealed that the C-terminal portion (residues 188 to 393, designated Lyr188-393) of Lyr was completely identical to the N-terminal portion (residues 1 to 206, designated NAGPR1-206) of the N-acetyl-γ-glutamyl-phosphate reductase (NAGPR) encoded by the E. coli K-12 argC gene (14). Also, the N-terminal region (residues 1 to 187, designated Lyr1-187) of Lyr was similar to the N-terminal region (residues 1 to 187, designated NAAAR1-187) of the N-acyl amino acid racemase (NAAAR) from Deinococcus radiodurans ATCC 13939 (18), with 98.4% identity. This led us to ascertain both the NAGPR and NAAAR activities of Lyr. As the amino acid sequence of residues 188 to 393 of Lyr showed 100% identity to the N-terminal portion of E. coli NAGPR, DNA fragments corresponding to these two coding regions were, respectively, cloned from the lyr and E. coli argC genes into pQE30 to generate plasmids plyr188-393 and pargC1-206. Since residues 1 to 187 of Lyr exhibited 98.4% identity to the residues in the N-terminal region of D. radiodurans NAAAR, DNA fragments corresponding to these two coding regions were also cloned from the lyr and naaar genes, respectively, to generate plasmids plyr1-187 and pnaaar1-187. Recombinant plasmids were then introduced into E. coli NovaBlue and induced by the addition of 0.5 mM IPTG and incubation at 30°C for 6 h. The recombinant proteins in the crude extract were purified by nickel chelate affinity chromatography. The activities of NAAAR and NAGPR were assayed by methods described previously (18, 34). The results showed that purified Lyr188-393 and NAGPR1-206 had about 50% of the specific activity of E. coli NAGPR, while the purified NAAAR1-187, Lyr, and Lyr1-187 proteins displayed no racemization activity toward chiral N-acetyl-d,l-methionine, N-acetyl-d,l-phenylalanine, N-acetyl-d,l-homophenyl-alanine, or N-d,l-carbamoyl-homophenylalanine (Table 2). Moreover, no lysine racemase activity could be detected from NAGPR, NAAAR, the truncated proteins, or an equimolar mixture of the Lyr1-187 and Lyr188-393 proteins.
TABLE 2.
Enzyme activities of various recombinant proteins
| Protein | Sp act (U/mg)
|
|
|---|---|---|
| Reductive dephosphorylation of N-acetyl-γ-glutamyl-phosphatea | Racemization of N-acetyl-d-methionineb | |
| NAGPR | 0.95 ± 0.014 | —c |
| NAGPR1-206 | 0.47 ± 0.017 | ND |
| Lyr | NDd | ND |
| Lyr188-393 | 0.46 ± 0.029 | ND |
| NAAAR | — | 1.91 ± 0.057 |
| NAAAR1-187 | — | ND |
| Lyr1-187 | — | ND |
The NAGPR activity assay.
The NAAAR activity assay.
—, not determined.
ND, not detectable.
The C-terminal portion of Lyr contained a region essential for NAGPR activity including the LIAVPGCYPTAAQLALKP domain (residues 335 to 352) as a catalytic region (3, 13), the GxxGxxG motifs (residues 195 to 201 and 372 to 377) as an NADP-binding region (24), and the ATAHEVSHDL region (residues 266 to 275) for Zn binding (24), which might explain the truncated protein Lyr188-393, with about 50% of the specific activity of the native NAGPR enzyme from E. coli. Nevertheless, these two truncated proteins lack racemization activity toward lysine. D. radiodurans NAAAR acts on a broad range of N-acyl amino acids rather than amino acids (38). The active-site framework for D. radiodurans NAAAR, comprising Lyr170, Asp195, Glu220, Asp245, and Lys269, which catalyze 1,1-proton exchange of N-acyl amino acids (38), was not present in the Lyr protein. This could also be proved by the fact that Lyr and the truncated proteins NAAAR1-187 and Lyr1-187 did not show racemization activity toward N-acetyl-d-methionine (Table 2). These findings support the view that the partial regions of NAAAR and NAGPR might have been combined to evolve a novel enzyme, Lyr, in which the new catalytic sites were created for catalyzing the racemization of lysine.
The identities of the N-terminal region (residues 1 to 187) and C-terminal region (residues 188 to 393) of Lyr to N-terminal regions of D. radiodurans NAAAR (residues 1 to 187) and E. coli NAGPR (residues 1 to 206), respectively, also suggest that Lyr can be regarded as a fusion protein of truncated NAAAR and NAGPR. Therefore, it could be speculated that DNA double-strand breaks (DSBs) of the genes for NAAAR and NAGPR might have occurred in bacterial host cells for the evolution of the lyr gene. In response to this situation, bacteria may use either homologous recombination or nonhomologous end joining (NHEJ) to repair DSBs in DNA (40). Since homologous DNA sequences at the ends of the N- and C-terminal regions of the lyr gene are not present to facilitate homologous recombination, NHEJ may be used to join broken DNA ends from the NAAAR- and NAGPR-encoding genes directly end to end (23). Recently, a DNA NHEJ complex has been identified in bacteria (40) and its role in repairing DSBs in the bacterial chromosome, especially in quiescent states such as sporulation or late stationary-phase culture, has been investigated (22, 26, 32, 37). The working model of bacterial NHEJ is that the end-binding protein Ku binds to the broken DNA ends, aligns them, and thus prepares for ligation, protects from degradation, and recruits DNA ligase LigD, which then directly catalyzes the sealing step of break repair (16). The LigD and Ku proteins are critical agents of the pathway which have not been found in the E. coli genome. Silicon sequence searches of sequenced bacterial genomes identified genes encoding putative NHEJ proteins in dozens of diverse bacterial genera, suggesting that NHEJ exists broadly in bacteria (8, 21, 41). Apparently, the gene for Lyr may have evolved from the truncated genes for NAAAR and NAGPR by NHEJ in bacterial cells other than those of E. coli. Since the N- and C-terminal regions of the lyr gene show about 98.4 and 98.7% DNA homology to the corresponding regions of the naaar and argC genes, in evolutionary terms, the lyr gene may have only recently evolved from E. coli NAGPR and D. radiodurans NAAAR.
Concluding remarks.
To the best of our knowledge, this is the first report of the cloning, purification, and characterization of a novel lyr gene from a soil microbial metagenome. Lyr can be regarded as a fusion protein of truncated NAAAR and NAGPR. Since NAAAR and NAGPR do not possess the activity needed to catalyze the racemization of lysine, a new catalytic site in Lyr should be determined in the future. Moreover, preliminary experiments in our laboratory showed that several plant species are sensitive to l-lysine, while d-lysine supports their growth. Based on these findings, we have successfully demonstrated the utility of the lyr gene as a novel selectable marker in the genetic transformation of tobacco and Arabidopsis with l-lysine as the selective agent (33). The studies also showed that the lyr gene is more efficient than the nptII gene traditionally used during the selection process in terms of normal growth of transgenic plants. One of the salient features of this gene is that it confers non-antibiotic-based resistance and therefore will meet the future requirement of using less controversial genes for the production of transgenic plants.
Nucleotide sequence accession number.
The nucleotide sequence of the lyr gene has been deposited in the GenBank database under accession number FJ405258.
Supplementary Material
Acknowledgments
This work was supported in part by a grant 93AS-4.1.2-FD-Z1-(1) from the Council of Agriculture, Taiwan.
Footnotes
Published ahead of print on 5 June 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, E. 1972. Amino acid racemases and epimerases, p. 479-507. In P. D. Boyer (ed.), The enzymes, vol. VI. Academic Press, New York, NY. [Google Scholar]
- 2.Amos, H., and G. N. Cohen. 1954. Amino acid utilization in bacterial growth. II. A study of threonine-isoleucine relationships in mutants of Escherichia coli. Biochem. J. 57:338-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco, J., R. A. Moore, and R. E. Viola. 2003. Capture of an intermediate in the catalytic cycle of l-aspartate-β-semialdehyde dehydrogenase. Proc. Natl. Acad. Sci. USA 100:12613-12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buschiazzo, A., M. Goytia, F. Schaeffer, W. Degrave, W. Shepard, C. Gregoire, N. Chamond, A. Cosson, A. Berneman, N. Coatnoan, P. M. Alzari, and P. Minoprio. 2006. Crystal structure, catalytic mechanism, and mitogenic properties of Trypanosoma cruzi proline racemase. Proc. Natl. Acad. Sci. USA 103:1705-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y. F., and E. Adams. 1974. d-Lysine catabolic pathway in Pseudomonas putida: interrelations with l-lysine catabolism. J. Bacteriol. 117:753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y. F., and E. Adams. 1977. Factors influencing growth on l-lysine by Pseudomonas. Regulation of terminal enzymes in the δ-aminovalerate pathway and growth stimulation by α-ketoglutarate. J. Biol. Chem. 252:7987-7991. [PubMed] [Google Scholar]
- 7.Chang, Y. F., and E. Adams. 1977. Glutarate semialdehyde dehydrogenase of Pseudomonas. Purification, properties, and relation to l-lysine catabolism. J. Biol. Chem. 252:7979-7986. [PubMed] [Google Scholar]
- 8.Cheng, C., and S. Shuman. 1997. Characterization of an ATP-dependent DNA ligase encoded by Haemophilus influenzae. Nucleic Acids Res. 25:1369-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, S. Y., N. Esaki, M. Ashiuchi, T. Yoshimura, and K. Soda. 1994. Bacterial glutamate racemase has high sequence similarity with myoglobins and forms an equimolar inactive complex with hemin. Proc. Natl. Acad. Sci. USA 91:10144-10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook, S. P., I. Galve-Roperh, A. Martinez del Pozo, and I. Rodriguez-Crespo. 2002. Direct calcium binding results in activation of brain serine racemase. J. Biol. Chem. 277:27782-27792. [DOI] [PubMed] [Google Scholar]
- 11.Corrigan, J. J. 1969. d-Amino acids in animals. Science 164:142-149. [DOI] [PubMed] [Google Scholar]
- 12.Fothergill, J. C., and J. R. Guest. 1977. Catabolism of l-lysine by Pseudomonas aeruginosa. J. Gen. Microbiol. 99:139-155. [DOI] [PubMed] [Google Scholar]
- 13.Gessert, S. F., J. H. Kim, F. E. Nargang, and R. L. Weiss. 1994. A polyprotein precursor of two mitochondrial enzymes in Neurospora crassa. Gene structure and precursor processing. J. Biol. Chem. 269:8189-8203. [PubMed] [Google Scholar]
- 14.Glansdorf, N., and G. Sand. 1965. Coordination of enzyme synthesis in the arginine pathway of Escherichia coli K-12. Biochim. Biophys. Acta 108:308-311. [DOI] [PubMed] [Google Scholar]
- 15.Glaser, L. 1960. Glutamic acid racemase from Lactobacillus arabinosus. J. Biol. Chem. 235:2095-2098. [PubMed] [Google Scholar]
- 16.Gong, C., P. Bongiorno, A. Martins, N. C. Stephanou, H. Zhu, S. Shuman, and M. S. Glickman. 2005. Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat. Struct. Mol. Biol. 12:304-312. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto, A., T. Nishikawa, T. Oka, K. Takahashi, and T. Hayashi. 1992. Determination of free amino acid enantiomers in rat brain and serum by high-performance liquid chromatography after derivatization with N-tert-butyloxycarbonyl-l-cysteine and o-phthaldialdehyde. J. Chromatogr. 582:41-48. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, S. K., H. H. Lo, W. D. Lin, I. C. Chen, C. H. Kao, and W. H. Hsu. 2007. Stereoselective synthesis of l-homophenylalanine using the carbamoylase method with in situ racemization via N-acylamino acid racemase. Process Biochem. 42:856-862. [Google Scholar]
- 19.Huang, H. T., and J. W. Davisson. 1958. Distribution of lysine racemase in bacteria. J. Bacteriol. 76:495-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichihara, A., and E. A. Ichihara. 1961. Metabolism of l-lysine by bacterial enzymes. V. Glutaric semialdehyde dehydrogenase. J. Biochem. 49:154-157. [DOI] [PubMed] [Google Scholar]
- 21.Magnet, S., and J. S. Blanchard. 2004. Mechanistic and kinetic study of the ATP-dependent DNA ligase of Neisseria meningitidis. Biochemistry 43:710-717. [DOI] [PubMed] [Google Scholar]
- 22.Moeller, R., E. Stackebrandt, G. Reitz, T. Berger, P. Rettberg, A. J. Doherty, G. Horneck, and W. L. Nicholson. 2007. Role of DNA repair by nonhomologous-end joining in Bacillus subtilis spore resistance to extreme dryness, mono- and polychromatic UV, and ionizing radiation. J. Bacteriol. 189:3306-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nonaka, T., A. Kita, J. Miura-Ohnuma, E. Katoh, N. Inagaki, T. Yamazaki, and K. Miki. 2005. Crystal structure of putative N-acetyl-γ-glutamyl-phosphate reductase (AK071544) from rice (Oryza sativa). Proteins 61:1137-1140. [DOI] [PubMed] [Google Scholar]
- 25.Peters, H. L., A. C. Davis, and B. T. Jones. 2004. Enantiomeric separations of amino acids with inductively coupled plasma carbon emission detection. Microchem. J. 76:85-89. [Google Scholar]
- 26.Pitcher, R. S., A. J. Green, A. Brzostek, M. Korycka-Machala, J. Dziadek, and A. J. Doherty. 2007. NHEJ protects mycobacteria in stationary phase against the harmful effects of desiccation. DNA Repair (Amsterdam) 6:1271-1276. [DOI] [PubMed] [Google Scholar]
- 27.Revelles, O., M. Espinosa-Urgel, T. Fuhrer, U. Sauer, and J. L. Ramos. 2005. Multiple and interconnected pathways for l-lysine catabolism in Pseudomonas putida KT2440. J. Bacteriol. 187:7500-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarower, M. G., T. Matsui, and H. Abe. 2003. Distribution and characteristics of d-amino acid and d-aspartate oxidases in fish tissues. J. Exp. Zool. A 295:151-159. [DOI] [PubMed] [Google Scholar]
- 29.Soda, K., and N. Esaki. 1994. Pyridoxal enzymes acting on d-amino acids. Pure Appl. Chem. 66:709-714. [Google Scholar]
- 30.Soda, K., T. Yorifuji, and K. Ogata. 1967. Occurrence of arginine racemase in bacterial extract. Biochim. Biophys. Acta 146:606-608. [DOI] [PubMed] [Google Scholar]
- 31.Stadtman, T. C., and P. Elliott. 1957. Studies on the enzymic reduction of amino acids. II. Purification and properties of d-proline reductase and a proline racemase from Clostridium sticklandii. J. Biol. Chem. 228:983-997. [PubMed] [Google Scholar]
- 32.Stephanou, N. C., F. Gao, P. Bongiorno, S. Ehrt, D. Schnappinger, S. Shuman, and M. S. Glickman. 2007. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J. Bacteriol. 189:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thiruvengadam, V., I. C. Chen, W. D. Lin, and W. H. Hsu. 2008. Lysine racemase: a novel selectable marker system for plant transformation, abstr. P-2-4, p. 96. In Abstracts of the 4th International Symposium on Biocatalysis and Biotechnology, Taipei, Taiwan.
- 34.Tokuyama, S., and K. Hatano. 1995. Cloning, DNA sequencing and heterologous expression of the gene for thermostable N-acylamino acid racemase from Amycolatopsis sp. TS-1-60 in Escherichia coli. Appl. Microbiol. Biotechnol. 42:884-889. [DOI] [PubMed] [Google Scholar]
- 35.Uo, T., T. Yoshimura, N. Tanaka, K. Takegawa, and N. Esaki. 2001. Functional characterization of alanine racemase from Schizosaccharomyces pombe: a eucaryotic counterpart to bacterial alanine racemase. J. Bacteriol. 183:2226-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh, C. T. 1989. Enzymes in the d-alanine branch of bacterial cell wall peptidoglycan assembly. J. Biol. Chem. 264:2393-2396. [PubMed] [Google Scholar]
- 37.Wang, S. T., B. Setlow, E. M. Conlon, J. L. Lyon, D. Imamura, T. Sato, P. Setlow, R. Losick, and P. Eichenberger. 2006. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 358:16-37. [DOI] [PubMed] [Google Scholar]
- 38.Wang, W. C., W. C. Chiu, S. K. Hsu, C. L. Wu, C. Y. Chen, J. S. Liu, and W. H. Hsu. 2004. Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans. J. Mol. Biol. 342:155-169. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe, T., K. Shibata, Y. Kera, and R. Yamada. 1998. Occurrence of free d-aspartate and aspartate racemase in the blood shell Scapharca broughtonii. Amino Acids 14:353-360. [DOI] [PubMed] [Google Scholar]
- 40.Weller, G. R., B. Kysela, R. Roy, L. M. Tonkin, E. Scanlan, M. Della, S. K. Devine, J. P. Day, A. Wilkinson, F. d'Adda di Fagagna, K. M. Devine, R. P. Bowater, P. A. Jeggo, S. P. Jackson, and A. J. Doherty. 2002. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 297:1686-1689. [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson, A., J. Day, and R. Bowater. 2001. Bacterial DNA ligases. Mol. Microbiol. 40:1241-1248. [DOI] [PubMed] [Google Scholar]
- 42.Wood, W. A., and I. C. Gunsalus. 1951. d-Alanine formation; a racemase in Streptococcus faecalis. J. Biol. Chem. 190:403-416. [PubMed] [Google Scholar]
- 43.Wurges, K., K. Petrusevska, S. Serci, S. Wilhelm, C. Wandrey, A. Seidel-Morgenstern, M. P. Elsner, and S. Lutz. 2009. Enzyme-assisted physiochemical enantioseparation processes. Part I. Production and characterization of a recombinant amino acid racemase. J. Mol. Catal. B 58:10-16. [Google Scholar]
- 44.Yamada, M., and K. Kurahashi. 1968. Adenosine triphosphate and pyrophosphate dependent phenylalanine racemase of Bacillus brevis Nagano. J. Biochem. 63:59-69. [DOI] [PubMed] [Google Scholar]
- 45.Yoshimura, T., and N. Esak. 2003. Amino acid racemases: functions and mechanisms. J. Biosci. Bioeng. 96:103-109. [DOI] [PubMed] [Google Scholar]
- 46.Yoshimura, T., N. Esaki, and K. Soda. 1992. Structure and function of alanine racemase. Bull. Inst. Chem. Res. Kyoto Univ. 70:378-384. [Google Scholar]
- 47.Zenk, M. H., and H. Scherf. 1963. d-Tryptophan in höheren Pflanzen. Biochim. Biophys. Acta 71:737-738. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.