Abstract
Photoheterotrophic microbes, which are capable of utilizing dissolved organic materials and harvesting light energy, include coccoid cyanobacteria (Synechococcus and Prochlorococcus), aerobic anoxygenic phototrophic (AAP) bacteria, and proteorhodopsin (PR)-containing bacteria. Our knowledge of photoheterotrophic microbes is largely incomplete, especially for high-latitude waters such as the Arctic Ocean, where photoheterotrophs may have special ecological relationships and distinct biogeochemical impacts due to extremes in day length and seasonal ice cover. These microbes were examined by epifluorescence microscopy, flow cytometry, and quantitative PCR (QPCR) assays for PR and a gene diagnostic of AAP bacteria (pufM). The abundance of AAP bacteria and PR-containing bacteria decreased from summer to winter, in parallel with a threefold decrease in the total prokaryotic community. In contrast, the abundance of Synechococcus organisms did not decrease in winter, suggesting that their growth was supported by organic substrates. Results from QPCR assays revealed no substantial shifts in the community structure of AAP bacteria and PR-containing bacteria. However, Arctic PR genes were different from those found at lower latitudes, and surprisingly, they were not similar to those in Antarctic coastal waters. Photoheterotrophic microbes appear to compete successfully with strict heterotrophs during winter darkness below the ice, but AAP bacteria and PR-containing bacteria do not behave as superior competitors during the summer.
Photoheterotrophy, which is the ability to utilize organic substrates and to harvest light energy, occurs in a broad range of microbes (14). Phototrophic microbes should be included in models of carbon cycling and food web dynamics, which now typically include only photoautotrophs, which produce organic carbon and oxygen, and heterotrophs, which consume organic matter and oxygen via aerobic respiration (55). Photoheterotrophy is potentially an important competitive adaptation, enabling microbes to survive adverse conditions or to outgrow competitors. Photoheterotrophic microbes include proteorhodopsin (PR)-containing bacteria, aerobic anoxygenic phototrophic (AAP) bacteria, and cyanobacteria.
PR is a membrane protein that binds retinal and functions as a light-driven proton pump that can have several physiological functions, including ATP generation (15). The actual role of PR in the environment is uncertain, however. Light enhances the growth of some PR-containing bacteria, such as Dokdonia sp. (17), but has no effect on the growth of others, including Pelagibacter ubique (16) and the SAR92-like strain HTCC2207 (44). Similarly, Campbell et al. (4) found no significant correlation with light intensity for three of four PR gene types examined in the North Atlantic Ocean. Nevertheless, emerging biogeographic patterns of PR genes are providing clues about what controls the distribution and abundance of PR-containing photoheterotrophs in oceanic systems. One of the first oceanic environments to be examined for PR was the coastal waters near Palmer Station, Antarctica (2). Sequence analysis revealed that the Antarctic PRs differed from those isolated from Monterey Bay and surface waters in the central North Pacific (2). In spite of this early report, there has been no work on PR-containing bacteria in Arctic waters. PR-containing bacteria may have unique responses to the continuous summer light, winter darkness, and shading by seasonal ice cover that occur in high-latitude environments.
The diversity and abundance of AAP bacteria have been examined by sequencing of the pufM gene (20, 51, 58), which is involved in bacteriochlorophyll (BChl a) synthesis, and by counting of BChl a-fluorescing cells by infrared fluorescence microscopy (14). Enumeration by infrared epifluorescence microscopy indicates that the abundance of AAP bacteria in environments such as the North Pacific Gyre and the Northeast Atlantic Ocean ranges from 1% to 10% (12, 13, 42) and can exceed 10% of the total prokaryotic community in estuaries (41, 50). AAP bacteria have been found in freshwater high-latitude waters (20, 35), but sequence analysis of pufM genes indicates that these AAP bacteria are distinct from those found in marine systems (50). The abundance of AAP bacteria decreases with latitude within the North Atlantic Ocean, from the central gyre to the waters near Greenland (13). Although these photoheterotrophic microbes are still present at 65°N, extrapolation of the trend suggests that AAP bacteria might be absent from the high-latitude waters of the Arctic Ocean.
Polar waters appear to be an exception to the otherwise widespread distribution of coccoid cyanobacteria in the world oceans (33, 54). The abundance of Synechococcus and Prochlorococcus decreases with latitude, as exemplified by the 4-orders-of-magnitude decline in abundance between 44°S and 62°S in the South Atlantic Ocean (25). The abundance of Synechococcus also decreases with latitude in the North Atlantic Ocean, between the central gyre and the waters near Greenland, to a low level at 65°N (13). The strong correlation between abundance and temperature (25) suggests that coccoid cyanobacteria are not important at high latitudes, although there are scattered reports of Prochlorococcus in waters as far north as 60°N, near Iceland (27), and of Synechococcus in Antarctic coastal waters (53). However, more data are needed on the abundance of Synechococcus and Prochlorococcus in polar waters such as the Arctic Ocean.
The goal of this study was to explore the abundance and diversity of photoheterotrophic microbes in the Arctic Ocean in order to develop a better picture of the biogeographic range of these biogeochemically important microbes and to gain insights into their ecology. Coastal waters of the Chukchi Sea and the Beaufort Sea were sampled in summer at the end of 24-h daylight and in winter following the period of 24-h darkness. The abundances of cyanobacteria, PR-containing bacteria, and AAP bacteria were monitored using flow cytometry, infrared epifluorescence microscopy, and real-time quantitative PCR (QPCR). These data provide a unique perspective on the potential impact of photoheterotrophic microbes on food webs and carbon cycling in this high-latitude aquatic system.
MATERIALS AND METHODS
Sample collection.
Seawater samples were collected from a small boat in the summer and through a hole in the ice in the winter. Seawater was pumped from a depth of 2 m into carboys and transported in insulated boxes back to the shore-based lab in Barrow, AK. Seawater was collected on 11 July 2008 from two locations in the Chukchi Sea, located at 71°25′21.60"N, 156°51′38.88"W and 71°23′4.80"N, 156°48′43.08"W. On 13 July, seawater was collected in the Beaufort Sea at two locations, 71°26′52.20"N, 156°3′22.80"W and 71°23′27.00"N, 156°9′3.24"W. In winter 2008, seawater was collected on 26 January at 71°21′9.90"N, 156°40′59.50"W, on 28 January at 71°20′39.90"N, 156°40′39.50"W, and on 30 January at 71°21′9.90"N, 156°40′39.50"W.
Biological oceanographic parameters.
Chl a was measured by fluorometry, using 90% acetone extracts of particulate material collected on GF/F (Whatman) filters (32). [3H]leucine incorporation rates were measured by standard methods (21). Triplicate samples with [3H]leucine (20 nmol liter−1) were incubated for 1.5 to 2.5 h at in situ temperatures of 4.0°C in summer and −1.8°C in winter. The incorporated [3H]leucine was precipitated by trichloroacetic acid (TCA), collected by centrifugation, and then rinsed with 5% TCA and 80% ethanol. The samples were then dried and radioassayed to determine [3H]leucine incorporation. Killed controls were poisoned with TCA.
Seawater samples for nutrient analyses were frozen and stored at −20°C until analysis. Concentrations of nitrate plus nitrite, phosphate, and silicate were determined by automated, segmented flow colorimetric analysis, using a Flo-Solution IV analyzer (O/I Analytical, College Station, TX). The concentration of NH4+ was determined using the sodium nitroprusside method (43) immediately upon returning the seawater to the Barrow lab.
Microscopic microbial enumeration.
Prokaryote abundance was determined using paraformaldehyde-fixed samples that were filtered onto 0.2-μm-pore-size black polycarbonate filters and then stained with a solution containing 1 μg ml−1 4′,6-diamidino-2-phenylindole (DAPI) (36) in a Tris buffer (pH 7.2) with 0.5 M NaCl. Total prokaryotes were enumerated by epifluorescence microscopy using semiautomated image analysis (11).
AAP bacteria were enumerated using an Intensified Retiga charge-coupled device camera (QImaging) mounted on a computer-controlled microscope (Olympus Provis AX70) with image analysis software (ImagePro Plus; Media Cybernetics) following the procedure described by Cottrell et al. (12). A series of four images was acquired for each field of view, using the following optical filter sets: DAPI (excitation wavelength, 360 ± 40 nm; emission wavelength, 460 ± 50 nm), infrared (390 ± 100 nm and 750-nm long pass), Chl a (480 ± 30 nm and 660 ± 50 nm), and phycoerythrin (545 ± 30 nm and 610 ± 75 nm) (Chroma). Cells were identified by detecting edges with Laplacian and Gaussian filters applied in series (26). The filtered images were segmented into binary format and then overlaid to identify AAP bacteria, which have DAPI and infrared fluorescence but not Chl a or phycoerythrin fluorescence.
Seawater samples for enumeration of coccoid cyanobacteria were preserved with 2% paraformaldehyde and stored at −20°C until analysis. Analyses were performed with a FACSCalibur (BD Biosciences) flow cytometer, using a 488-nm laser excitation wavelength and 0.2-μm-pore-size filtered sheath fluid prepared with deionized water and 35 g liter−1 NaCl. Synechococcus spp. were identified in plots of red (>640 nm) versus orange (560 to 640 nm) fluorescence (5). Counts were calibrated using flow rates determined by weighing samples before and after analysis. Consistency of flow rates, fluorescence, and side scatter was monitored using 1-μm-diameter fluorescent beads (Molecular Probes) added to samples.
Nucleic acid extraction.
Seawater was prefiltered through 0.8-μm polycarbonate filters to minimize the abundance of eukaryotes. Cells in the bacterial size fraction were collected on 0.22-μm Durapore (Millipore) filters by vacuum filtration, and the filters were stored at −80°C in a cetyltrimethyl ammonium bromide buffer (9). Nucleic acids were extracted using standard protocols (3), and the DNA concentration was determined using a Pico Green assay (Invitrogen, Carlsbad, CA).
Diversity in pufM and PR genes.
The diversity in pufM and PR genes was estimated using clone libraries of PCR amplicons. Established procedures were used to amplify pufM genes, using PCR primers pufM557F and pufM_WAWR (50), and PR genes were amplified using PCR primers PR-1aF and PR-1aR (4). Briefly, PCR amplification used the following conditions: 1 cycle of 94°C for 2 min and 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 30 s. Final concentrations of primers and MgCl2 were 1.87 mM and 2 mM, respectively. PCR products were cloned with a Topo TA cloning kit for sequencing (Invitrogen) according to the manufacturer's instructions. Colonies were screened via PCR to determine the insert size, and 96 clones of the expected size were sequenced by standard Sanger sequencing. Sequences were trimmed to remove any vector and PCR primer sequences and aligned using the CLUSTAL program in ARB (23). Representative sequences most closely related to the PR clone sequences were retrieved from GenBank and added to the alignment. The Arctic pufM sequences were aligned to the sequences analyzed by Yutin et al. (59). Genetic distances were calculated using ARB for analysis by S-Libshuff (39).
Quantification of pufM and PR gene abundance.
The abundance of pufM and PR genes in Chukchi Sea and Beaufort Sea seawater samples was determined by real-time QPCR. The abundances of summer and winter clades of pufM and PR genes were determined using QPCR primers targeting these groups (Table 1). QPCR primers were designed from the nucleotide sequences of our Arctic clones. The abundance of bacterial 16S rRNA genes was determined using the QPCR primers and amplification conditions described by Suzuki et al. (47) in order to calculate the abundance of pufM and PR genes relative to the abundance of bacterial 16S rRNA genes. Standard curves were generated using plasmid clones in the QPCR assays of pufM and PR genes. Plasmid DNA was linearized with the PstI restriction enzyme and quantified using the Pico Green assay (Invitrogen). Standard curves for QPCR analysis of bacterial 16S rRNA genes were generated using Escherichia coli genomic DNA.
TABLE 1.
PCR primers and conditions used in QPCR analysis of PR, pufM, and 16S rRNA gene abundances
| Gene | Group | Target region | Primer name | Primer sequence (5′-3′) | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|---|
| 16S rRNA gene | Bacteria | 1369-1541a | BACT1369F | CGGTGAATACGTTCYCGG | 58 | 47 |
| PROK1541R | AAGGAGGTGATCCRGCCGCA | |||||
| PR gene | Summer clade | 472-567b | PRS472F | ATCGTAGGCATGGTAGGC | 55 | This study |
| PRS567R | CTTGACAGATTCTGGAGC | |||||
| Winter clade | 460-562b | PRW460F | ACCGTGGCCTTCATCATT | 60 | This study | |
| PRW562R | CTGATGGGGGTGCGCTTT | |||||
| pufM | Summer clade | 657-760c | pufMS621F | CATGCATGGTGCCACGAT | 55 | This study |
| pufMS724R | AGAAGAGAGCGGCACGTT | |||||
| Winter clade | 562-641c | pufMW526F | ACGTCGGCCATCTCCCTG | 55 | This study | |
| pufMW622R | TGGCAAACAGCAGCACAG |
Nucleotide positions in the E. coli 16S rRNA gene.
Nucleotide positions in the Pelagibacter ubique PR gene.
Nucleotide positions in the Roseobacter sp. strain SO3 (GenBank accession no. AY675566) pufM gene.
QPCR was performed in triplicate, using 1 μl of environmental sample DNA or plasmid DNA solution and a Taq polymerase mixture containing Sybr green (Stratagene) in a total reaction volume of 12.5 μl. Thermal cycling and quantification were performed using an ABI 7500 instrument programmed with the following thermal cycling conditions: 95°C for 10 min followed by 30 to 40 cycles of denaturation at 95°C for 15 s, primer annealing at the primer-specific annealing temperature (Table 1) for 45 s, and polymerase extension at 72°C for 45 s. The final primer concentration was 0.2 mM. Amplification efficiency averaged 99.2% ± 0.4%, and amplification specificity was always achieved, as determined by the presence of a single peak in each postamplification dissociation curve.
Nucleotide sequence accession numbers.
The nucleotide sequences of the Arctic pufM and PR genes were deposited in GenBank under accession numbers FJ618945 to FJ619035.
RESULTS
The abundance of photosynthetic biomass and Chl-containing picoeukaryotes and the concentration of inorganic nutrients varied between summer and winter in the Alaska coastal waters we sampled, reflecting the cessation of primary production during winter darkness. The standing stock of photosynthetic biomass, as determined by the concentration of Chl a, decreased substantially (eightfold) between summer and winter (Table 2). Similarly, flow cytometric analysis revealed that the abundance of Chl a-containing picoeukaryotes decreased 200-fold, from 5.4 × 106 cells/liter in the summer to 0.02 × 106 cells/liter in the winter (Table 2). The decrease in photosynthetic biomass was accompanied by 3- to 20-fold increases from summer to winter in the concentrations of nitrate plus nitrate, phosphate, ammonium, and silicate (Table 2).
TABLE 2.
Chl a concentrations, picoeukaryote abundances, and inorganic nutrient concentrations in Arctic coastal waters in summer 2007 and winter 2008a
| Season | Chl a concn (μg/liter) | Concn of picoeukaryotes (106 cells/liter) | Nitrate-plus-nitrite concn (μM) | Phosphate concn (μM) | Ammonium concn (μM) | Silicate concn (μM) | Temp (°C) | Salinity (psu) |
|---|---|---|---|---|---|---|---|---|
| Summer 2007 | 0.48 ± 0.11 | 5.37 ± 1.83 | 0.57 ± 0.4 | 0.38 ± 0.18 | 0.32 ± 0.08 | 7.28 ± 3.1 | 4.1 ± 1.3 | 27 ± 6 |
| Winter 2008 | 0.06 ± 0.01 | 0.02 ± 0.01 | 11.3 ± 0.19 | 1.95 ± 0.08 | 1.88 ± 0.23 | 22.9 ± 0.85 | −1.8 ± 0 | 35 ± 0.5 |
Data are means ± standard deviations. The data represent values measured at four sites in the Beaufort Sea and the Chukchi Sea in summer 2007 and at three sites in the Chukchi Sea in winter 2008. psu, practical salinity units.
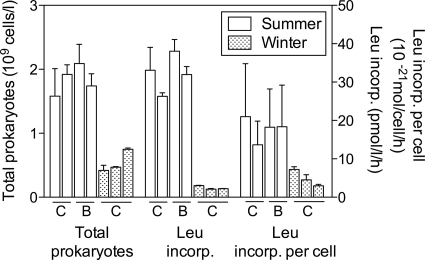
Prokaryotic abundance decreased between summer and winter, but the change was not as great as that of autotrophic biomass. The abundance of prokaryotes decreased threefold, from almost 2 × 109 cells/liter in July to about 0.5 × 109 cells/liter in January (Fig. 1). Incorporation of [3H]leucine decreased 13-fold, much more than the abundance of prokaryotes. Leucine incorporation per cell decreased fourfold, from 17.7 amol leucine/cell/h in the summer to 4.5 amol leucine/cell/h in the winter (Fig. 1).
FIG. 1.
Abundance of total prokaryotic cells and rates of [3H]leucine incorporation in the coastal Chukchi Sea (C) and Beaufort Sea (B) in summer 2007 and winter 2008. Values are shown for four sites sampled in summer and three sites sampled in winter.
Abundance of Synechococcus and AAP bacteria.
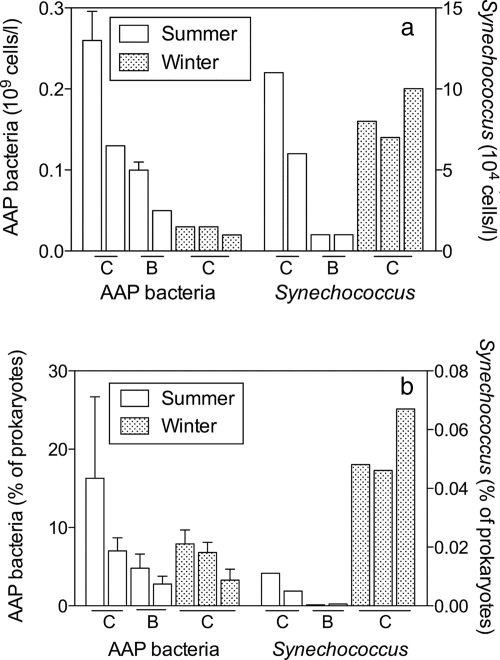
Synechococcus cells were detected in every sample collected during the summer and, surprisingly, during the winter as well. Prochlorococcus was never detected. Synechococcus abundance averaged 4 × 104 to 8 × 104 cells/liter and did not differ significantly between winter and summer, although there was much variation between sampling sites, especially in the summer (Fig. 2a). Consequently, because total prokaryote abundance decreased, the percentage of Synechococcus organisms increased 15-fold, from 0.004% in the summer to 0.05% in the winter (Fig. 2b).
FIG. 2.
Abundance (a) and contribution to total prokaryotic abundance (b) of Synechococcus and AAP bacteria in the coastal Chukchi Sea (C) and Beaufort Sea (B) in summer 2007 and winter 2008. Values are shown for four sites sampled in summer and three sites sampled in winter.
AAP bacteria were 1,000-fold more abundant than Synechococcus organisms in winter and in summer. The abundance of AAP bacteria decreased fourfold between the seasons, from 0.13 × 109 cells/liter in the summer to 0.03 × 109 cells/ml in the winter (Fig. 2a). However, the contribution of AAP bacteria to the total prokaryotic community did not change between the two seasons. In both seasons, AAP bacteria made up 5 to 8% of the prokaryotes (Fig. 2b).
Arctic pufM genes.
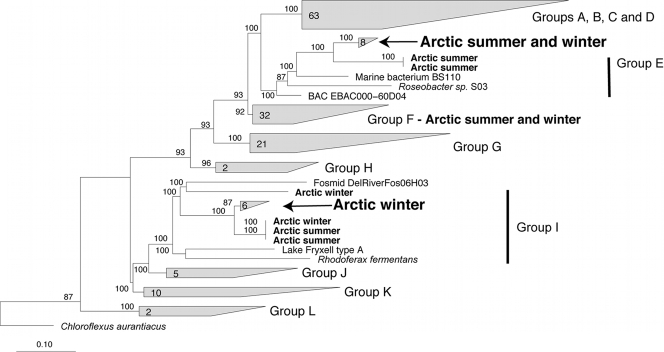
Phylogenetic analysis of pufM genes amplified from Arctic environmental DNA revealed sequences belonging to clades previously identified in a biogeographic survey of these genes (59). All of the Arctic pufM genes could be assigned to one of the 12 groups of pufM genes identified by Yutin et al. (59), including groups E, F, and I (Fig. 3). Most of the Arctic pufM genes belonged to group F, which did not vary in relative abundance between summer and winter (Table 3). In contrast, the second most abundant group differed between the summer and winter samples. In the summer, 31% of the pufM genes belonged to group I, and in the winter, group E accounted for 38% of these genes. The least abundant groups also differed between seasons. Groups E and I accounted for 4% and 12% of the summer and winter sequences, respectively (Table 3).
FIG. 3.
Phylogenetic tree of pufM genes, including Arctic pufM genes amplified by PCR from the coastal Chukchi Sea and Beaufort Sea in summer 2007 and winter 2008. Arctic sequences, which are indicated in bold, were added to the pufM gene tree previously described by Yutin et al. (59), using the ARB quick-add tool. The clades labeled groups A to L were identified in the previous analysis of Yutin et al. (59). The arrows indicate Arctic clades of pufM genes that were targeted by QPCR. The numbers of sequences within the clades are indicated at the bases of the trapezoids, and bootstrap percentages appear at the branch nodes.
TABLE 3.
Composition of pufM gene clone libraries constructed in summer and in wintera
| Group | % Similarity (mean ± SD) | n | % of pufM clones
|
|
|---|---|---|---|---|
| Summer | Winter | |||
| E | 95 ± 4 | 10 | 4 | 38 |
| F | 99 ± 1 | 27 | 65 | 50 |
| I | 96 ± 2 | 9 | 31 | 12 |
According to the classification proposed by Yutin et al. (59).
The S-Libshuff test (39) indicated no significant difference between the two clone libraries of pufM genes obtained in the winter and summer. However, phylogenetic analysis revealed clades dominated by summer and winter sequences (Fig. 3). Clades dominated by summer and winter sequences were identified in groups E and F, respectively. Group I contained a clade that included a mixture of summer and winter sequences. Within the summer and winter clades, nucleotide and amino acid similarities were >96% and 94%, respectively (Table 4).
TABLE 4.
Abundances and similarities of clones in libraries of Arctic pufM and PR genes from summer and wintera
| Gene | Clade | No. of clones
|
% Similarity
|
||
|---|---|---|---|---|---|
| Summer library | Winter library | Nucleotide | Amino acid | ||
| pufM | Summer | 7 | 1 | 89 | 96 |
| Winter | 0 | 6 | 97 | 100 | |
| Total | 24 | 23 | |||
| PR gene | Summer | 9 | 0 | 97 | 98 |
| Winter | 0 | 6 | 95 | 98 | |
| Total | 20 | 24 | |||
The summer and winter clades were chosen for monitoring by QPCR.
PCR primers were designed to target the summer and winter clades of pufM genes (Table 1). The primers targeting the summer clade matched every sequence in the summer clade and had at least five mismatches to sequences outside the summer clade, except for two closely related sequences in group E that were also retrieved from the summer sample (Fig. 3). Similarly, the primers targeting the winter clade matched every sequence in the winter clade and had more than six mismatches to other Arctic sequences, except for three closely related winter sequences in group I.
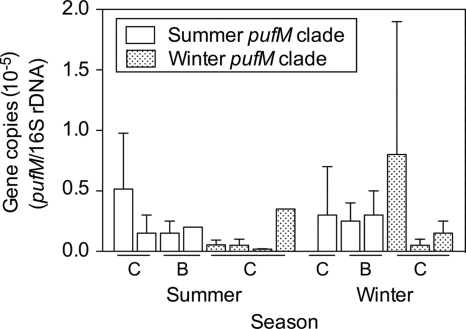
There was no consistent seasonal change in the abundance of pufM genes belonging to the summer and winter clades (t test; P > 0.05). The abundance of the summer pufM gene clade was about 0.5 × 10−5 gene copies per 16S rRNA gene in both summer and winter (Fig. 4). Similarly, there was no significant difference in the abundance of the winter pufM gene cluster between summer and winter (t test; P > 0.05). The abundance of the winter pufM gene clade was 0.2 × 10−5 ± 0.3 × 10−5 gene copies per 16S rRNA gene in summer and 0.7 × 10−5 ± 0.8 × 10−5 gene copies per 16S rRNA gene in winter (Fig. 4). The errors reported here represent spatial variation among the four locations sampled in summer and the three locations sampled in winter. Even though the average abundance of the winter pufM gene clade was >3-fold higher in winter than in summer, the variation between sampling sites masked any statistical significance.
FIG. 4.
Abundance of Arctic pufM genes relative to the abundance of bacterial 16S rRNA genes in the coastal Chukchi Sea (C) and Beaufort Sea (B) in summer 2007 and winter 2008. Clades of pufM genes identified in summer and winter samples were targeted using clade-specific QPCR primers. Values are shown for four sites sampled in summer and three sites sampled in winter.
Arctic PR genes.
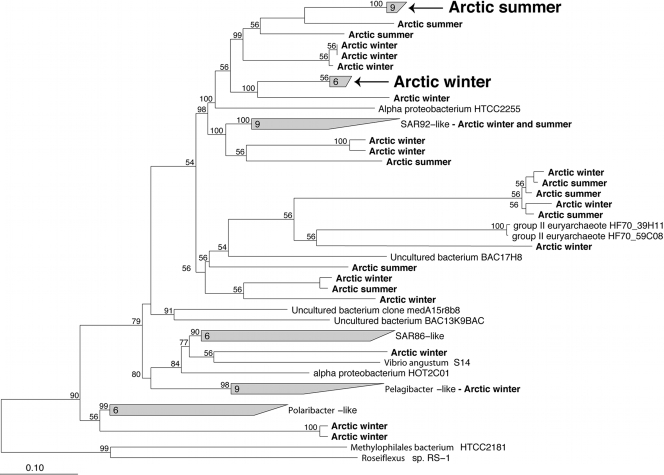
Phylogenetic analysis revealed a diverse collection of Arctic PR genes, but only a few sequences belonged to previously identified groups (Fig. 5). For example, only one Arctic PR gene from the winter sequences clustered together with the PR genes from Pelagibacter-like bacteria. None of the Arctic PR genes clustered together with Polaribacter-like PR genes, SAR86-like PR genes, or genes of the group II-like euryarchaeote group. The only previously recognized cluster containing Arctic genes was the SAR92-like group, which had six Arctic genes, with three each from the summer and winter samples (Fig. 5).
FIG. 5.
Phylogenetic tree of PR genes amplified by PCR from the coastal Chukchi Sea and Beaufort Sea in summer 2007 and winter 2008. The ARB quick-add tool was used to add the Arctic sequences, which are indicated in bold, to a tree of complete PR gene sequences constructed using Bayesian estimation of phylogeny (1, 38). The arrows indicate clades of Arctic pufM genes identified in summer and winter samples targeted by QPCR. The numbers of sequences in the clades are indicated at the bases of the trapezoids, and the bootstrap percentages appear at the nodes.
Two clades of PR genes included sequences found only in the summer or winter samples. The summer clade included nine PR genes, and the winter clade included six genes (Fig. 5). The summer and winter clones were >95% and 98% similar, respectively, at the DNA level and >98% similar at the protein level (Table 4). There was no obvious seasonal clustering of the other Arctic PR genes, which belonged to clades (supported by a bootstrap value of 56%) containing two to five PR genes. The clades containing mixtures of PR genes from summer and winter were made up of highly similar PR genes as well. For example, the three summer and three winter PR genes comprising a clade adjacent to the group II archaeal PR genes were >96% similar at the DNA and protein levels.
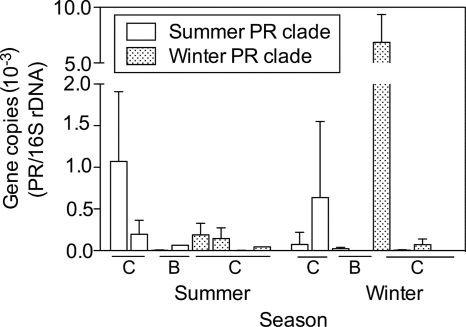
There was no consistent seasonal shift in the abundance of PR genes belonging to the summer and winter clades (Fig. 6). The abundance of PR genes in either the summer or winter clade did not differ between summer and winter (t test; P > 0.05), but the winter clade was about 10-fold more abundant than the summer clade, although the difference was not significant because of variation between sampling sites. The abundance of the PR genes belonging to the summer clade averaged 3 × 10−4 PR gene copies per 16S rRNA gene in both seasons, while the winter clade's abundance was 1 × 10−3 to 2 × 10−3 copies per 16S rRNA gene (Fig. 6).
FIG. 6.
Abundance of Arctic PR genes relative to the abundance of bacterial 16S rRNA genes in the coastal Chukchi Sea (C) and Beaufort Sea (B) in summer 2007 and winter 2008. Clades of PR genes identified in summer and winter samples were targeted using clade-specific QPCR primers. Values are shown for four sites sampled in summer and three sites sampled in winter. Note the break in the y axis.
DISCUSSION
The main objectives of this study were to examine photoheterotrophic microbes in the Arctic Ocean and to assess changes in their abundance between summer and winter. Extremes in day length and seasonal ice cover might be expected to result in unique impacts of photoheterotrophic microbes on carbon cycling in high-latitude waters. In addition, the ability to consume dissolved organic materials (DOM) and to harvest light energy could provide photoheterotrophic microbes with a competitive advantage that varies seasonally with light availability. Sampling after continuous summer sunlight and after winter darkness constituted a natural experiment for examining the responses of photoheterotrophs to light and dark in the Arctic Ocean. All three groups of photoheterotrophs examined here were present in coastal waters of the Chukchi Sea and the Beaufort Sea in both summer and winter. Seasonal shifts in the abundances of PR-containing bacteria and AAP bacteria between summer and winter paralleled changes in the abundance of the total prokaryotic community. As a result, the abundance of PR-containing bacteria and AAP bacteria remained a constant percentage of total prokaryote abundance between summer and winter.
The absence of a seasonal shift in the relative abundance of PR and pufM genes belonging to summer and winter clades was unexpected. We anticipated that clades representing the most successful microbes capable of utilizing sunlight would dominate the PCR clone libraries in the summer. However, QPCR analysis revealed no seasonal shift in the abundance of these microbes relative to the total prokaryotic community. These data might suggest that light harvesting does not provide photoheterotrophs with a superior advantage in summer. Of course, there may be problems in inferring abundances from clone libraries of PCR products because of possible biases in the PCR step (45, 46), as seen for 16S rRNA genes (10). Similarly, QPCR analysis has potential biases, including problems with extraction efficiencies, unknown gene copy numbers, and genome sizes that would interfere with absolute quantification (47). However, these problems were avoided by determining the abundances of PR and pufM genes relative to that of 16S rRNA genes (47).
The PR and pufM genes identified in the Arctic Ocean are phylogenetically distinct from genes seen in lower-latitude waters. The most similar lower-latitude PR gene, which was from a Sargasso Sea metagenome (49), was only 78% similar to the Arctic gene at the DNA level. The Arctic pufM genes were also different from those in Antarctic Lake Fryxell (20). The highest similarity between Arctic and Lake Fryxell pufM genes was 83%. Unfortunately, there are no pufM gene sequences available from the Southern Ocean or Antarctic coastal waters. The Arctic PR genes were also distinct from those found at lower latitudes, and surprisingly, they were not at all similar to those found in Antarctic coastal waters (2). The Antarctic sequences all belonged to the SAR86 clade of PR genes, but none of the Arctic PR genes belonged to this group, and they were at most 58% similar to those found in Antarctic waters. PR genes appear to differ between the Northern and Southern Hemispheres, and in contrast to the case for pufM, there do not appear to be strictly polar communities of PR genes.
It is unclear how AAP bacteria generate BChl a in the summer under continuous light. High levels of irradiance block BChl a synthesis entirely in cultivated AAP bacteria (57), and exposure to even low light downregulates pigment synthesis (22). Those AAP bacterial strains capable of generating BChl a under continuous illumination produce substantial quantities of the pigment only under microaerophilic conditions (28). It appears that light effects on pigment synthesis are different in high-latitude environments than those seen in culture. Inhibition of BChl a synthesis could lead to high pigment content in the winter and low pigment content in the summer. Such shifts in pigment content could lead to underestimates of AAP bacterial abundance in the summer if the level of pigment fluorescence fell below the microscopic detection level. However, estimating abundance by microscopy did not appear to have been confounded by a low pigment content per cell. The QPCR analysis of pufM genes produced the same pattern in the abundance of AAP bacteria between summer and winter as that seen for data from epifluorescence microscopy.
The variation in Synechococcus abundance between summer and winter was quite different from that seen for PR-containing bacteria and AAP bacteria. Synechococcus abundance in the winter was equal to the summer abundance, which suggests that Synechococcus sustains sufficient growth throughout the winter to balance mortality. In contrast, total prokaryotic abundance decreased significantly in winter, together with a fourfold decrease in growth rate; doubling times based on [3H]leucine incorporation averaged 33 ± 6 days in summer and 133 ± 57 days in winter. It seems unlikely that the constancy of Synechococcus abundance reflects escape from mortality in the winter, because prokaryotes are actively grazed during winter in the Arctic, when bacterivory appears to control bacterial production (48). In addition, cyanophages infecting Synechococcus are prevalent in the Arctic waters of the Chukchi Sea and the Beaufort Sea (7).
The fact that Synechococcus organisms are present in the Arctic Ocean during winter may not be too surprising because consumption of DOM is now well recognized as an important feature of cyanobacterial metabolism in the ocean. Analysis of a fully sequenced Synechococcus genome revealed genes homologous to those encoding transport systems for the uptake of organic substrates such as amino acids, oligopeptides, and cyanate (31). Similarly, uptake experiments indicated that organic sulfur compounds provide a significant amount of S to Synechococcus and that Synechococcus spp. are important consumers of dimethylsulfoniopropionate in the ocean (24). Prochlorococcus and Synechococcus appear to be responsible for much of the light effect on leucine incorporation in oceanic waters (8, 27). Our data on Synechococcus suggest that DOM consumption in the dark may be important to the survival of Synechococcus during winter darkness and a potentially important feature of carbon cycling in the Arctic.
The presence of Synechococcus spp. in Arctic Ocean waters greatly extends the known range of these cyanobacteria, which previously were believed to be limited to lower latitudes by water temperature (29, 34). In fact, Synechococcus has been used to distinguish between warm Atlantic waters and cold Arctic waters (18, 19, 30). Our data indicate that low temperatures, which averaged 4.1°C in summer and −1.8°C in winter, did not restrict Synechococcus from the Chukchi Sea and the Beaufort Sea. Previous reports hinted at a potential role for Synechococcus in high-latitude waters. For example, Synechococcus strains have been seen in some lakes and coastal seas of Antarctica (37). However, our results are the first to demonstrate the potential importance of Synechococcus in the Arctic Ocean and to provide clues about adaptation to low temperatures and long periods of winter darkness.
In addition to in situ growth, there are several potential allochthonous sources of Synechococcus cells. Walker and Marchant (53) used ice dynamics to explain the summer and winter peaks in Synechococcus abundance recorded near Davis Station, Antarctica. Ice scouring potentially resuspends cells from the benthos into the water column in winter, and ice melting could release cells trapped in sea ice itself in the summer (53). Synechococcus abundance may depend on freshwater delivery to the Arctic Ocean. The Mackenzie River is an important source of cyanobacteria in the eastern Beaufort Sea (Northwest Territories, Canada), but the input of Synechococcus from the river decreases 10-fold 200 km offshore (52). However, riverine inputs are not a likely source of Synechococcus in our study region because no major rivers discharge into the region of the Chukchi Sea and the western Beaufort Sea that we sampled.
We also considered the possibility that photoheterotrophs found in Arctic coastal waters are transported from the Pacific Ocean by advection. Pacific Ocean water flows into the Arctic Ocean and can be identified by its distinct salinity signature (56). Although meteorological forcing by prevailing northerly winds may have a strong influence on the southerly movement of water masses in the Chukchi Sea (56), the net flow is to the north, transporting Pacific Ocean water from the Bering Strait to the Arctic Ocean (6). The transport level of Pacific Ocean water is large (56) and has a substantial impact on the salinity of the Arctic Ocean. The importance of advection for transporting microbes to our coastal sites is unclear. Current meter studies indicate that the annual mean current velocity in the central Chukchi Sea is about 5 cm/s, which yields a 4-month transit time from the Bering Strait to Barrow Canyon (56). This transit time is similar to in situ microbial turnover times, which are on the order of 4 months in the winter but 30 days in the summer.
The results of this study have several implications for understanding photoheterotrophy and photoheterotrophic microbes in the ocean. Extended periods of winter darkness did not stop photoheterotrophic microbes from inhabiting these polar waters. Nevertheless, the benefit of light harvesting is apparently not sufficient to enable blooms of photoheterotrophic microbes in the summer. Incubation experiments conducted with coastal Pacific Ocean water also failed to detect shifts in PR-containing bacteria and AAP bacteria, but those experiments lasted only 5 to 10 days (40). Even though our sampling was conducted after months of continuous light and darkness, there was still no indication that light enables PR-containing bacteria or AAP bacteria to flourish in the Arctic when sunlight is available. Similarly, there seems to be no competitive disadvantage to PR-containing bacteria and AAP bacteria when sunlight is no longer available in winter. Winter abundances of Synechococcus equivalent to summer levels suggest that mortality is balanced by metabolism supported by DOM consumption. Targeted studies of DOM consumption by photoheterotrophs will further elucidate their roles in carbon cycling in the Arctic Ocean.
Acknowledgments
This work was supported by an NSF grant (OPP0632233).
We thank Glenn Christman for his help with field sampling. Glenn Sheehan and Lewis Brower provided logistics support at the Barrow Arctic Science Consortium.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Altekar, G., S. Dwarkadas, J. P. Huelsenbeck, and F. Ronquist. 2004. Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20:407-415. [DOI] [PubMed] [Google Scholar]
- 2.Béjà, O., E. N. Spudich, J. L. Spudich, M. Leclerc, and E. F. DeLong. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786-789. [DOI] [PubMed] [Google Scholar]
- 3.Bostrom, K. H., K. Simu, A. Hagström, and L. Riemann. 2004. Optimization of DNA extraction for quantitative marine bacterioplankton community analysis. Limnol. Oceanogr. Methods 2:365-373. [Google Scholar]
- 4.Campbell, B. J., L. A. Waidner, M. T. Cottrell, and D. L. Kirchman. 2008. Abundant proteorhodopsin genes in the North Atlantic Ocean. Environ. Microbiol. 10:99-109. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, L. 2001. Flow cytometric analysis of autotrophic picoplankton. Methods Microbiol. 30:317-343. [Google Scholar]
- 6.Carmack, E., D. Barber, J. Christensen, R. Macdonald, B. Rudels, and E. Sakshaug. 2006. Climate variability and physical forcing of the food webs and the carbon budget on panarctic shelves. Prog. Oceanogr. 71:145-181. [Google Scholar]
- 7.Chenard, C., and C. A. Suttle. 2008. Phylogenetic diversity of sequences of cyanophage photosynthetic gene psbA in marine and freshwaters. Appl. Environ. Microbiol. 74:5317-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Church, M. J., H. W. Ducklow, and D. A. Karl. 2004. Light dependence of [3H]leucine incorporation in the oligotrophic North Pacific Ocean. Appl. Environ. Microbiol. 70:4079-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke, J. D. 2009. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harbor Protoc. 2009:5177-5178. [DOI] [PubMed] [Google Scholar]
- 10.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 12.Cottrell, M. T., A. Mannino, and D. L. Kirchman. 2006. Aerobic anoxygenic phototrophic bacteria in the Mid-Atlantic Bight and the North Pacific Gyre. Appl. Environ. Microbiol. 72:557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cottrell, M. T., V. K. Michelou, N. Nemcek, G. DiTullio, and D. L. Kirchman. 2008. Carbon cycling by microbes influenced by light in the northeast Atlantic Ocean. Aquat. Microb. Ecol. 50:239-250. [Google Scholar]
- 14.Eiler, A. 2006. Evidence for the ubiquity of mixotrophic bacteria in the upper ocean: implications and consequences. Appl. Environ. Microbiol. 72:7431-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuhrman, J. A., M. S. Schwalbach, and U. Stingl. 2008. Proteorhodopsins: an array of physiological roles? Nat. Rev. Microbiol. 6:488-494. [DOI] [PubMed] [Google Scholar]
- 16.Giovannoni, S. J., L. Bibbs, J. C. Cho, M. D. Stapels, R. Desiderio, K. L. Vergin, M. S. Rappé, S. Laney, L. J. Wilhelm, H. J. Tripp, E. J. Mathur, and D. F. Barofsky. 2005. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438:82-85. [DOI] [PubMed] [Google Scholar]
- 17.Gómez-Consarnau, L., J. M. González, M. Coll-Lladó, P. Gourdon, T. Pascher, R. Neutze, C. Pedrós-Alió, and J. Pinhassi. 2007. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature 445:210-213. [DOI] [PubMed] [Google Scholar]
- 18.Gradinger, R., and J. Lenz. 1989. Picocyanobacteria in the high Arctic. Mar. Ecol. Progr. Ser. 52:99-101. [Google Scholar]
- 19.Gradinger, R., and J. Lenz. 1995. Seasonal occurrence of picocyanobacteria in the Greenland Sea and central Arctic Ocean. Polar Biol. 15:447-452. [Google Scholar]
- 20.Karr, E. A., W. M. Sattley, D. O. Jung, M. T. Madigan, and L. A. Achenbach. 2003. Remarkable diversity of phototrophic purple bacteria in a permanently frozen Antarctic lake. Appl. Environ. Microbiol. 69:4910-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchman, D. L. 2001. Measuring bacterial biomass production and growth rates from leucine incorporation in natural aquatic environments. Methods Microbiol. 30:227-237. [Google Scholar]
- 22.Li, Q., N. Z. Jiao, and Z. Q. Peng. 2006. Environmental control of growth and BChl a expression in an aerobic anoxygenic phototrophic bacterium, Erythrobacter longus (DSMZ6997). Acta Oceanol. Sin. 25:138-144. [Google Scholar]
- 23.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malmstrom, R. R., R. P. Kiene, M. Vila, and D. L. Kirchman. 2005. Dimethylsulfoniopropionate (DMSP) assimilation by Synechococcus in the Gulf of Mexico and northwest Atlantic Ocean. Limnol. Oceanogr. 50:1924-1931. [Google Scholar]
- 25.Marchant, H. J., A. T. Davidson, and S. W. Wright. 1987. The distribution and abundance of chroococcoid cyanobacteria in the Southern Ocean. Proc. Natl. Inst. Polar Res. Symp. Polar Biol. 1:1-19. [Google Scholar]
- 26.Massana, R., J. M. Gasol, P. K. Bjornsen, N. Blackburn, A. Hagström, S. Hietanen, B. H. Hygum, J. Kuparinen, and C. Pedrós-Alió. 1997. Measurement of bacterial size via image analysis of epifluorescence preparations: description of an inexpensive system and solutions to some of the most common problems. Sci. Mar. 61:397-407. [Google Scholar]
- 27.Michelou, V. K., M. T. Cottrell, and D. L. Kirchman. 2007. Light-stimulated bacterial production and amino acid assimilation by cyanobacteria and other microbes in the North Atlantic Ocean. Appl. Environ. Microbiol. 73:5539-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montecchia, M. S., N. L. Pucheu, N. L. Kerber, and A. F. Garcia. 2006. Oxygen and light effects on the expression of the photosynthetic apparatus in Bradyrhizobium sp. C7T1 strain. Photosynth. Res. 90:215-222. [DOI] [PubMed] [Google Scholar]
- 29.Murphy, L. S., and E. M. Haugen. 1985. The distribution and abundance of phototrophic ultraplankton in the North Atlantic. Limnol. Oceanogr. 30:47-58. [Google Scholar]
- 30.Not, F., R. Massana, M. Latasa, D. Marie, C. Colson, W. Eikrem, C. Pedros-Alio, D. Vaulot, and N. Simon. 2005. Late summer community composition and abundance of photosynthetic picoeukaryotes in Norwegian and Barents Seas. Limnol. Oceanogr. 50:1677-1686. [Google Scholar]
- 31.Palenik, B., B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb, and J. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424:1037-1042. [DOI] [PubMed] [Google Scholar]
- 32.Parsons, T. R., Y. Maita, and C. M. Lalli. 1984. A manual of chemical and biological methods for seawater analysis, 1st ed. Pergamon Press, New York, NY.
- 33.Partensky, F., J. Blanchot, and D. Vaulot. 1999. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review, p. 457-475. In L. Charpy and A. W. D. Larkum (ed.), Marine cyanobacteria, vol. 19. Musée Océanographique, Monaco. [Google Scholar]
- 34.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perreault, N. N., C. W. Greer, D. T. Andersen, S. Tille, G. Lacrampe-Couloume, B. S. Lollar, and L. G. Whyte. 2008. Heterotrophic and autotrophic microbial populations in cold perennial springs of the high Arctic. Appl. Environ. Microbiol. 74:6898-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 37.Powell, L. M., J. P. Bowman, J. H. Skerratt, P. D. Franzmann, and H. R. Burton. 2005. Ecology of a novel Synechococcus clade occurring in dense populations in saline Antarctic lakes. Mar. Ecol. Prog. Ser. 291:65-80. [Google Scholar]
- 38.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 39.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwalbach, M. S., M. Brown, and J. A. Fuhrman. 2005. Impact of light on marine bacterioplankton community structure. Aquat. Microb. Ecol. 39:235-245. [Google Scholar]
- 41.Schwalbach, M. S., and J. A. Fuhrman. 2005. Wide-ranging abundances of aerobic anoxygenic phototrophic bacteria in the world ocean revealed by epifluorescence microscopy and quantitative PCR. Limnol. Oceanogr. 50:620-628. [Google Scholar]
- 42.Sieracki, M. E., I. C. Gilg, E. C. Thier, N. J. Poulton, and R. Goericke. 2006. Distribution of planktonic aerobic anoxygenic photoheterotrophic bacteria in the northwest Atlantic. Limnol. Oceanogr. 51:38-46. [Google Scholar]
- 43.Solorzano, L. 1969. Determination of ammonia in natural waters by the phenolhypochlorite method. Limnol. Oceanogr. 14:799-801. [Google Scholar]
- 44.Stingl, U., R. A. Desiderio, J. C. Cho, K. L. Vergin, and S. J. Giovannoni. 2007. The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl. Environ. Microbiol. 73:2290-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki, M., M. S. Rappé, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaqué, D., S. Guadayol, F. Peters, J. Felipe, L. Angel-Ripoll, R. Terrado, C. Lovejoy, and C. Pedrós-Alió. 2008. Seasonal changes in planktonic bacterivory rates under the ice-covered coastal Arctic Ocean. Limnol. Oceanogr. 53:2427-2438. [Google Scholar]
- 49.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Y. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 50.Waidner, L. A., and D. L. Kirchman. 2007. Aerobic anoxygenic phototrophic bacteria attached to particles in turbid waters of the Delaware and Chesapeake Estuaries. Appl. Environ. Microbiol. 73:3936-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waidner, L. A., and D. L. Kirchman. 2008. Diversity and distribution of ecotypes of the aerobic anoxygenic phototrophy gene pufM in the Delaware estuary. Appl. Environ. Microbiol. 74:4012-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waleron, M., K. Waleron, W. F. Vincent, and A. Wilmotte. 2007. Allochthonous inputs of riverine picocyanobacteria to coastal waters in the Arctic Ocean. FEMS Microbiol. Ecol. 59:356-365. [DOI] [PubMed] [Google Scholar]
- 53.Walker, T. D., and H. J. Marchant. 1989. The seasonal occurrence of chroococcoid cyanobacteria at an Antarctic Coastal Site. Polar Biol. 9:193-196. [Google Scholar]
- 54.Waterbury, J. B., S. W. Watson, F. W. Valois, and D. G. Franks. 1986. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus, p. 71-120. In T. Platt and W. K. W. Li (ed.), Photosynthetic picoplankton. Department of Fisheries and Oceans, Ottawa, Ontario, Canada.
- 55.Williams, P. J. L. 2000. Heterotrophic bacteria and the dynamics of dissolved organic material, p. 153-200. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, NY.
- 56.Woodgate, R. A., K. Aagaard, and T. J. Weingartner. 2005. A year in the physical oceanography of the Chukchi Sea: moored measurements from autumn 1990-1991. Deep-Sea Res. II 52:3116-3149. [Google Scholar]
- 57.Yurkov, V. V., and J. T. Beatty. 1998. Aerobic anoxygenic phototrophic bacteria. Microbiol. Mol. Biol. Rev. 62:695-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yutin, N., O. Béjà, and M. Suzuki. 2008. The use of denaturing gradient gel electrophoresis with fully degenerate pufM primers to monitor aerobic anoxygenic phototrophic assemblages. Limnol. Oceanogr. Methods 6:427-440. [Google Scholar]
- 59.Yutin, N., M. T. Suzuki, H. Teeling, M. Weber, J. C. Venter, D. B. Rusch, and O. Béjà. 2007. Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling Expedition metagenomes. Environ. Microbiol. 9:1464-1475. [DOI] [PubMed] [Google Scholar]