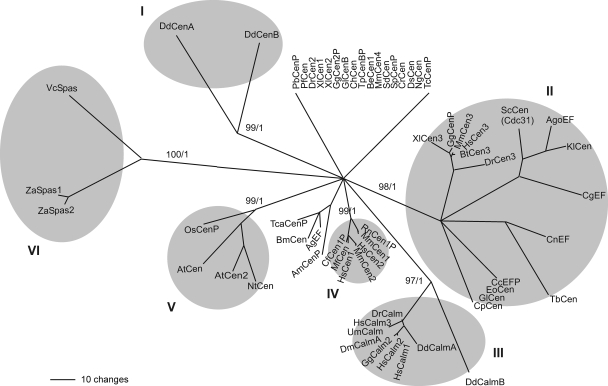
FIG. 2.
Phylogenetic analysis shows that DdCenB, together with DdCenA, constitutes a divergent clade within the EF-hand superfamily of proteins (I). The main clades are shaded, and the bootstrap (top) and Bayesian support values (bottom) are shown for each one of them. Resolved sequences in this phylogram were present in at least 80% of the trees generated by Bootstrap, otherwise they are shown as unresolved. Nonconserved sequence flanking the four EF-hand domains at the N and C termini of all proteins were disregarded in this analysis. Cen, centrin; Calm, calmodulin; Spas, spasmin; EF, EF-hand-containing protein. Putative proteins were labeled with a “P” at the end of the name. GI, gene identification as annotated at NCBI (http://www.ncbi.nlm.nih.gov/sites/entrez?db=Protein&itool=toolbar). The phylogenetic relationships were estimated by the maximum parsimony analysis using PAUP* v. 4.0b. Bayesian analysis was performed with MrBayes 3.1.2 using eight chains for 1,100,000 generations, sampling every one thousand trees and with a burn-in of 100 trees. AgEF, Anopheles gambiae STR.PEST (GI: 116117847); AgoEF, Ashbya gossypii (GI: 45184714); AmCenP, Apis mellifera (GI: 66529823); AtCen2, Arabidopsis thaliana (GI: 15229732); AtCen, Arabidopsis thaliana (GI: 7270650); BeCen1, Blastocladiella emersonii (GI: 70931046); BmCen, Bombyx mori (GI: 87248293); BtCen3, Bos taurus (GI: 111307090); CcEFP, Coprinopsis cinerea okayama 7#130 (GI: 116509538); CfCen1P, Canis lupus familiaris (GI: 57089853); CgEF, Candida glabrata (GI: 50292355); ChCen, Cryptosporidium hominis TU502 (GI: 54658657); CnEF, Cryptococcus neoformans var. neoformans JEC21 (GI: 58270616); CpCen, Cryptosporidium parvum (GI: 46227249); CrCen, Chlamydomonas reinhardtii (GI: 159482892); DdCalmA, Dictyostelium discoideum AX4 (GI: 66815357); DdCalmB, Dictyostelium discoideum AX4 (GI: 66825411); DdCenA, Dictyostelium discoideum (GI: 66806051); DdCenB, Dictyostelium discoideum (GI: 66815669); DmCalmA, Drosophila melanogaster (GI: 17647231); DrCalm, Danio rerio (GI: 41054633); DrCen2, Danio rerio (GI: 189540405); DrCen3, Danio rerio (GI: 66472718); DsCen, Dunaliella salina (GI: 1705641); EoCen, Euplotes octocarinatus (GI: 75029523); GgCalm2, Gallus gallus (GI: 45384366); GgCen2P, Gallus gallus (GI: 50745880); GgCenP, Gallus gallus (GI: 118104383); GlCen, Giardia lamblia ATCC 50803 (GI: 159110457); GlCenB, Giardia lamblia ATCC 50803 (GI: 159114706); HsCalm1, Homo sapiens (GI: 5901912); HsCalm2, Homo sapiens (GI: 13623675); HsCalm3, Homo sapiens (GI: 13477325); HsCen1, Homo sapiens (GI: 4757974); HsCen2, Homo sapiens (GI: 4757902); HsCen3, Homo sapiens (GI: 46397403); KlCen, Kluyveromyces lactis (GI: 50305339); MfCen, Macaca fascicularis (GI: 67971808); MmCen1, Mus musculus (GI: 76253942); MmCen2, Mus musculus (GI: 10257421); MmCen3, Mus musculus (GI: 6680922); MmCen4, Mus musculus (GI: 22003866); NgCen, Naegleria gruberi (GI: 1705642); NtCen, Nicotiana tabacum (GI: 6358509); OsCenP, Oryza sativa (Japonica cultivar-group) (GI: 78708509); PbCenP, Plasmodium berghei (GI: 56499292); PfCen, Plasmodium falciparum 3D7 (GI: 124505775); RnCen1P, Rattus norvegicus (GI: 34877910); ScCen (Cdc31), Saccharomyces cerevisiae (GI: 6324831); SdCen, Scherffelia dubia (GI: 21209); SpCenP, Strongylocentrotus purpuratus (GI: 115898527); TbCen, Trypanosoma brucei TREU927 (GI: 72387904); TcaCenP, Tribolium castaneum (GI: 91081379); TcCenP, Trypanosoma cruzi (GI: 70886637); TpCenBP, Theileria parva strain Muguga (GI: 71027987); UmCalm, Ustilago maydis 521 (GI: 46099694); VcSpas, Vorticella convallaria (GI: 4100824); XlCen1, Xenopus laevis (GI: 1017791); XlCen2, Xenopus laevis (GI: 32766515); XlCen3, Xenopus laevis (GI: 11119117); ZaSpas1, Zoothamnium arbuscula (GI: 26453345); ZaSpas2, Zoothamnium arbuscula (GI: 26453347).