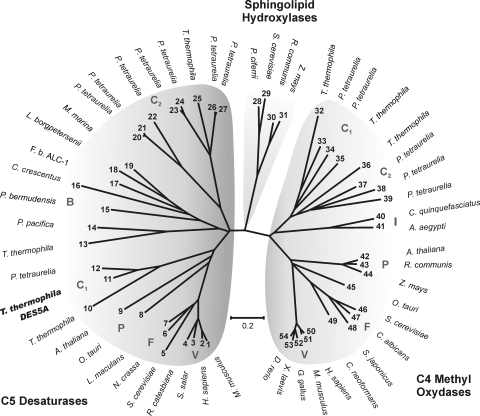
FIG. 6.
Phylogenetic analysis of C-5(6) sterol desaturases, C-4 sphingolipid hydroxylases, sterol C-4 methyl oxidases, and putative proteins of Tetrahymena thermophila. The phylogenetic tree was created using the neighbor-joining method, with 10,000 replicates, in MEGA-4 (24). The accession numbers of the amino acid sequences used for the analysis were the following: 1, NP_766357; 2, NP_008849; 3, NP_001133588; 4, ACO51759; 5, NP_013157; 6, XP_962923; 7, AAN27998; 8, CAL53849; 9, NP_186907; 10, XP_001023372 (TTHERM_00446080); 11, XP_001029976 (TTHERM_01194720); 12, XP_001440490; 13, XP_001017777 (TTHERM_00438800); 14, ZP_01908611; 15, ZP_01017596; 16, NP_420481; 17, ZP_02181983; 18, YP_798571; 19, ZP_01689977; 20, XP_001441873; 21, XP_001453241; 22, XP_001441700; 23, XP_001450297; 24, XP_001453136; 25, XP_001016978 (TTHERM_00758950); 26, XP_001426093; 27, XP_001459383; 28, AAN77731; 29, EDN60625; 30, EEF39915; 31, NP_001149259; 32, XP_001015720 (TTHERM_00077800); 33, XP_001449651; 34, XP_001455915; 35, XP_001022978 (TTHERM_00348230); 36, XP_001016047 (TTHERM_00876970); 37, XP_001448034; 38, XP_001460322; 39, XP_001459715; 40, XP_001861819; 41, XP_001657694; 42, NP_850133; 43, EEF41918; 44, ACG34890; 45, CAL54207; 46, NP_011574; 47, XP_713456; 48, XP_002174187; 49, XP_569526; 50, NP_006736; 51, NP_079712; 52, XP_420391; 53, NP_001072809; and 54, NP_998518. Specific branches are indicated as V (vertebrates), F (fungi), P (plant), B (bacteria), I (insect), and C1 and C2 (ciliates). The bar shows the percentages of substitutions.