Abstract
The human pathogen Cryptococcus neoformans causes meningoencephalitis. The polysaccharide capsule is one of the main virulence factors and consists of two distinct polysaccharides, glucuronoxylomannan (GXM) and galactoxylomannan (GalXM). How capsular polysaccharides are synthesized, transported, and assembled is largely unknown. Previously, it was shown that mutations in the CAP10, CAP59, CAP60, and CAP64 genes result in an acapsular phenotype. Here, it is shown that these acapsular mutants do secrete GalXM and GXM-like polymers. GXM and GalXM antibodies specifically reacted with whole cells and the growth medium of the wild type and CAP mutants, indicating that the capsule polysaccharides adhere to the cell wall and are shed into the environment. These polysaccharides were purified from the medium, either with or without anion-exchange chromatography. Monosaccharide analysis of polysaccharide fractions by gas-liquid chromatography/mass spectrometry showed that wild-type cells secrete both GalXM and GXM. The CAP mutants, on the other hand, were shown to secrete GalXM and GXM-like polymers. Notably, the GalXM polymers were shown to contain glucuronic acid. One-dimensional 1H nuclear magnetic resonance confirmed that the CAP mutants secrete GalXM and also showed the presence of O-acetylated polymers. This is the first time it is shown that CAP mutants secrete GXM-like polymers in addition to GalXM. The small amount of this GXM-like polymer, 1 to 5% of the total amount of secreted polysaccharides, may explain the acapsular phenotype.
Cryptococcus neoformans of the A (var. grubii [24]) and D (var. neoformans [36]) serotypes are the causative agents of cryptococcosis, of which the most common clinical form is meningoencephalitis. This disease is related to immunocompromised patients but can also occur in immunocompetent individuals (4, 19, 38). One of the main virulence factors is the polysaccharide capsule (2, 5, 17, 21, 27, 35). This capsule enables the yeast-like fungus to survive the harsh environment of the human body by using its immunomodulatory properties that enable immune evasion and by preventing killing through phagocytosis by macrophages (44, 45).
The capsule consists of a low percentage of mannoproteins (46) and the polysaccharides glucuronoxylomannan (GXM) and galactoxylomannan (GalXM) in a mass ratio of about 10:1 (14, 16, 17). Little is known about the synthesis of GXM and GalXM and their transport toward the cell surface. A mutation in the Sec4/Rab8 GTPase homologue was recently shown to affect protein secretion as well as polysaccharide secretion and resulted in intracellular accumulation of vesicles containing GXM (51). From this and the fact that GXM has been detected in extracellular vesicles, it was proposed that polysaccharides are packaged in such vesicles to cross the cell wall to reach the extracellular environment (47).
Mutation analysis has revealed four genes, called CAP10, CAP59, CAP60, and CAP64, which give an acapsular phenotype when inactivated (7, 9-13). The precise role of the encoded Cap proteins is unknown. Cap59 has been suggested to play a role in extracellular trafficking of multimeric forms of GXM molecules (26). Moreover, it may play a role in the assembly of GXM, since it shares homology with a mannosyltransferase (48). Like Cap59, Cap60 is a putative mannosyltransferase. Cap10 shares homology with a xylosyltransferase and therefore may also be involved in capsule assembly (34), like the recently identified xylosyltransferase encoded by CXT1 (33). This transferase has been shown to play a direct role in the synthesis of both of the capsular polysaccharides but is especially active in the addition of xyloses to the GalXM polysaccharide. CAP64 shares homology with so-called CAS genes, encoding proteins involved in O acetylation of GXM (40).
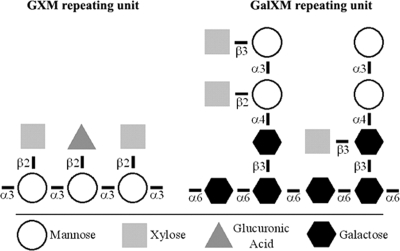
Structural analysis has revealed a relatively clear picture of the buildup of the GXM and GalXM polysaccharides (14, 50) (Fig. 1). Some variability in the chemical structures of the capsular polysaccharides has been described, even within the capsule of a particular strain (40, 50). In addition, GalXM has been shown to also contain, besides galactopyranose, galactofuranose in trace amounts (1, 29). The two C. neoformans serotypes A and D are distinguished based on variation in the position of the different xylose residues in the GXM repeating unit (30). The structure of the GalXM repeating unit was analyzed by using a fraction of purified polysaccharides secreted in the medium by a mutant of the D serotype called the CAP67 mutant. This strain is mutated in the same gene as a serotype A CAP59 mutant. The number of xylose residues can vary from zero up to six within the GalXM repeating unit (Fig. 1) (50).
FIG. 1.
Chemical structure of GXM and GalXM monomers. Large strands of these monomers form polymers of up to 1 × 106 to 7 × 106 daltons for GXM and 1 × 105 daltons for GalXM. Ratios vary between serotypes. Shown are serotype A GXM, Man 3/Xyl 2/GlcA 1, and GalXM, Gal 6/Man 4/Xyl 1.6 (shown are three xyloses). The degree of O acetylation is not shown. The picture is based on data from reference 3.
So far, secreted polysaccharides in the medium of the serotype D CAP67 mutant and the corresponding serotype A CAP59 mutant have been analyzed (41, 50). It was shown that these mutants secrete GalXM but not GXM in the medium. However, it is shown here that these mutants, as well as the serotype A CAP10, CAP60, and CAP64 mutants, also secrete GXM-like polymers in addition to GalXM. Moreover, part of GalXM seems to contain glucuronic acid, supporting earlier findings (16, 49).
MATERIALS AND METHODS
Strains and purified polysaccharides.
C. neoformans serotype A strain H99 (KNH99), its four isogenic derivatives with deletions in the CAP10 (NE305), CAP59 (NE367), CAP60 (NE309), and CAP64 (NE333) genes that were recently generated in the serotype A H99 background (42), and the serotype D CAP67 mutant (ATCC 52817) were used in this study. Saccharomyces cerevisiae NMY3Z was used for antibody preadsorption. In some of the experiments, GXM and GalXM, purified via the Cherniak et al. protocols (15, 20), were used. GXM and GalXM were purified from the C. neoformans serotype A strain ATCC 62066 (22) and the acapsular serotype D CAP67 mutant, respectively (20).
Growth conditions.
C. neoformans and S. cerevisiae were routinely grown at 200 rpm and 30°C in yeast-peptone-dextrose (2% [wt/vol] peptone, 1% [wt/vol] yeast extract, 2% [wt/vol] dextrose). Capsule production was induced under iron-limiting conditions in glucose-salts-urea (GSU) medium (2% [vol/vol] glucose, 21.5 mM urea, 10 mM KH2PO4, 1.2 mM MgSO4·7H2O, 1 μM MnCl2·4H2O, 10 μM FeSO4·7H2O, 10 μM ZnSO4·7H2O, 1.2 μM CuSO4·5H2O, 100 μM CaCl2·2H2O, 10 μg/liter biotin, and 2 mg/liter thiamine) (16).
Antibodies against capsule polysaccharides.
The polyclonal antisera anti-H99 and anti-GalXM have been raised in rabbits against heat-inactivated cells from C. neoformans strain H99 (a generous gift from Frank Coenjaerts, UMC, The Netherlands) and GalGlcXM (galactoglucoxylomannan) from Cryptococcus laurentii (6). Antisera were preincubated three times with 109 cells of S. cerevisiae NMY3Z for 1 h at room temperature to remove specific antibodies that could potentially bind yeast cell wall components. GXM-specific monoclonal antibodies 18B7 (1.9 mg/ml) (52) and 13F1 (1.4 mg/ml) (43) were a generous gift from the Casadevall lab (Albert Einstein College of Medicine, NY).
Staining of the C. neoformans capsule and cell wall.
A total of 107 to 109 cells were mixed at a dilution of 1:1 with a 1:1 dilution mix of India ink (8 12079 02249; Royal Talens) and Uvitex 2B (10 mg/ml). A total of 5 μl of the mixture was spotted on a slide and analyzed under a microscope (Olympus) with a 400× magnification. Photographs were made with the Nikon DX1200 digital camera and the Nikon ACT-1 software package (version 2.63). UV light was used to detect Uvitex-stained cells.
PI staining.
To determine the ratio of living cells to dead cells, cells were stained with propidium iodine (PI). Cryptococcus neoformans was grown in GSU medium at 30°C, and at different time intervals, aliquots were removed and stained with PI (5 μg/ml). Cells were analyzed under a microscope (Olympus) with a 400× magnification using 535 nm absorbance and 617 nm emission to detect PI-stained, dead cells.
Polysaccharide isolation.
Cells were grown in 50 ml GSU for 5 days, reaching a cell density of approximately 1 × 109 cells/ml in the case of the CAP10 mutant and 5 × 109 cells/ml for H99 and the other CAP mutants. The complete culture was subsequently used for polysaccharide purification. Cells were collected by centrifugation for 15 min at 10,400 × g. The supernatant was filtered over a 0.22-μm filter (SIGP033RS; Millipore), and the eluate was concentrated to 100 to 300 μl with a Centricon Plus-20 column (cutoff, 30 kDa; Millipore), according to the manufacturer's manual. Before use, the column was rinsed with deionized water. The sample was treated with DNase (2 units of DNase I) for 15 min at 37°C and with proteinase K (50 μg/ml) for 1 h at 37°C. After these treatments, 5% (wt/wt) trichloroacetic acid was added, and the mixture was incubated on ice for 15 min, followed by centrifugation at 20,800 × g for 15 min at 4°C. The supernatant, containing the polysaccharides, was extensively dialyzed against water (cutoff, 12 to 14 kDa, and the dialysis membrane was boiled for 10 min in water prior to use; Spectra/Por) and lyophilized. This isolation procedure yielded 100 to 300 mg of polysaccharide material from wild-type cells and 10 to 30 mg of polysaccharides from CAP mutant cells that were purified from 50-ml cultures in GSU medium.
Dot blotting.
Immunoblotting experiments for detection of polysaccharides in cells or in solution were performed, as described previously (31). More precisely, the indicated amounts of cells or polysaccharides extracted from the medium of wild-type and CAP mutant cells were spotted onto a nitrocellulose membrane and dried for 1 min with a blow-dryer. The membrane was blocked for 1 h in Tris-buffered saline (TBS; 10 mM Tris base, 150 mM NaCl [pH 7.5]) containing 1% Protifar or 1% gelatin (when the anti-GalXM antisera were used) and 0.2% Tween 20 and then incubated for 1 h at room temperature with either anti-H99 (1:200 dilution), anti-GalXM (1:200), 18B7 (1:200; final protein content, 9.7 μg/ml) or 13F1 (1:200; final protein content, 7 μg/ml) in TBS with 0.05% Tween 20 and 0.1% Protifar or 0.1% gelatin. Membranes were washed twice in TBS with 0.05% Tween 20 for 10 min and incubated for 1 h with either goat anti-rabbit immunoglobulin G (IgG)-alkaline phosphatase (1:10,000, ALI4405; Biosource) when the anti-H99 or anti-GalXM antisera were used or goat anti-mouse IgG-alkaline phosphatase (1:10,000, AMI4405; Biosource) when the 18B7 or 13F1 monoclonal antibodies were used. The membrane was washed once in TBS with 0.05% Tween 20, once in TBS, and once in phosphatase buffer (12.1 g Tris, 5.8 g NaCl, 1.0 g MgCl2·6H2O [pH 9.5]) for 10 min. Substrate was added (10 ml phosphatase buffer containing 50 μl Nitro Blue Tetrazolium chloride [NBT; 25 mg NBT dissolved in 1 ml of 70% dimethylformamide]) and 50 μl 5-bromo-4-chloro-3-indolylphosphate (BCIP; 10 mg BCIP dissolved in 1 ml dimethylformamide) until a signal appeared. The reaction was stopped with 1 mM EDTA, followed by storage at 4°C in water.
Immunofluorescence on cryptococcal cells.
A total of 108 cells were washed twice with PBS, resuspended in 750 μl PBS with 1% bovine serum albumin and incubated for 1 h at room temperature with either anti-H99 (1:200 dilution), anti-GalXM (1:200), 18B7 (9.7 μg/ml), or 13F1 (7 μg/ml). Cells were washed three times for 5 min in PBS with 1% bovine serum albumin and incubated with Alexa Fluor 594 goat anti-mouse IgG-conjugated antibodies (1:200 dilution, A11032; Molecular Probes) when the 18B7 or 13F1 monoclonal antibodies were used. Alternatively, Alexa Fluor 488 goat anti-rabbit IgG-conjugated antibodies (1:200 dilution, A11034; Molecular Probes) were used with the anti-H99 or anti-GalXM polyclonal antisera. After this, cells were incubated for 1 h at room temperature with gentle mixing and washed three times for 5 min with PBS. Cells were resuspended in 40 μl PBS and 10 μl Uvitex 2B (10 mg/ml), which binds to chitin in the cell wall (37). A total of 5 μl of the mixture was spotted on a slide and analyzed under a fluorescence microscope (Olympus) with 400× magnification. Photographs were made with the Nikon DX1200 digital camera and the Nikon ACT-1 software package (version 2.63), with identical exposure times and settings for each series of samples.
Anion-exchange chromatography.
A total of 0.5 to 5 mg of purified polysaccharide was loaded onto a 1-ml Q-Sepharose column (17-1014-01; Amersham Pharmacia), prewashed with 5 ml 1 M NaCl and 10 ml demi water. The column was eluted with 5 ml demieralized water, 5 ml 100 mM NaCl, 5 ml 250 mM NaCl, and 5 ml 1 M NaCl. Fractions were dialyzed against water (cutoff, 12 to 14 kDa; Spectra/Por) and lyophilized.
Monosaccharide analysis.
To estimate qualitatively the amount of polysaccharides in each fraction used for monosaccharide analysis, isolated polysaccharides in different dilutions were spotted onto a TLC plate (1.05735.0001; Merck) and stained with orcinol-sulfuric acid (100 mg orcinol, 95 ml methanol, 5 ml concentrated sulfuric acid [98%]) using hot air. Glucose was used as a standard. Hereafter, fractions were subjected to methanolysis (methanolic 1 M HCl for 24 h at 85°C) after addition of 50 to 100 nmol mannitol as an internal standard. The resulting mixture of (methyl ester) methyl glycosides was trimethylsilylated (hexamethyldisilazane-trimethylchlorosilane-pyridine dilution, 1:1:5; 30 min at room temperature) and analyzed by quantitative gas-liquid chromatography (GLC) with flame ionization detection and GLC/mass spectrometry (GLC MS). Quantitative GLC analysis was performed on a Chrompack CP9002 gas chromatograph, equipped with an EC-1 column (30 m by 0.32 mm; Alltech). GLC MS was carried out on a GC8060/MD800 system (Fisons Instruments, Interscience) (electron impact, 70 eV), using an AT-1 column (30 m by 0.25 mm; Alltech). In both cases, a temperature program of 140 to 240°C at 4°C/min was used. Glucuronic acid is known to be underestimated in these types of experiments, and therefore a correction factor was determined by using the tetrasaccharide Man(α1-3)[GlcA(β1-2)]Man(α1-3)Man as a reference compound. The amount of glucuronic acid detected in a known amount of tetrasaccharide was determined under similar experimental conditions by GLC MS. The amount of glucuronic acid is 2.5 times smaller than the expected value, giving a correction factor of 2.5 for the amount of glucuronic acid detected in the polysaccharide fractions.
The composition of the polysaccharide fractions was derived from the molar ratios of the monosaccharides obtained with quantitative GLC analysis. For purified GXM, the amount of mannose was set at three, which is the number of mannose residues in the repeating unit of GXM (14), and for purified GalXM, the amount of galactose was set at six, which is the number of galactopyranose residues in the repeating unit of GalXM (50). The amounts of the other monosaccharides were related to mannose or galactose.
NMR spectroscopy.
One-dimensional 500-MHz 1H nuclear magnetic resonance (NMR) spectra were recorded on a Bruker DRX-500 spectrometer, with a probe temperature of 300 K. Prior to analysis, samples were exchanged twice in D2O with intermediate lyophilization and then dissolved in 0.6 ml D2O. Chemical shifts (δ) are expressed in parts per million by reference to the monodeuterated water line (δ, 4.76; acetone at δ, 2.225).
RESULTS
Analysis of capsular components secreted by CAP mutants.
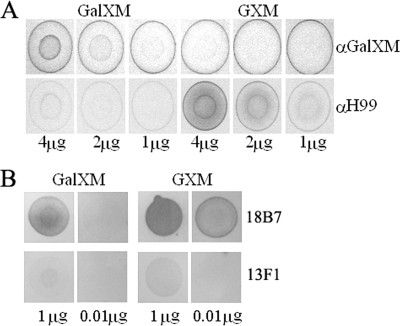
Mutations in the four CAP genes result in an acapsular phenotype, as determined by negative staining with India ink (data not shown). Fluorescence and dot blot immunolabeling was used to assess whether small amounts of capsule polysaccharides are still associated with the cell surface. To this end, the polyclonal antisera anti-H99 and anti-GalXM and the monoclonal antibodies 18B7 and 13F1 were used. The polyclonal serum anti-GalXM was raised in rabbits using purified polysaccharide GalGluXM (galactoglucoxylomannan) from Cryptococcus laurentii that was conjugated to a carrier protein (human serum albumin) (6). We anticipated and indeed found a cross-reactivity of this serum with the GalXM of Cryptococcus neoformans, as determined in dot immunoblotting experiments (Fig. 2A). The polyclonal antisera anti-H99 and anti-GalXM react mainly with GXM and GalXM, respectively, but particularly, the anti-H99 antiserum has cross-reactivity with purified GalXM (Fig. 2A). The monoclonal antibodies react specifically with GXM (52), but remarkably, we also detected reactivity with components present in the purified GalXM fraction in the dot blot assay (Fig. 2B). Remarkably, this GalXM fraction was derived from a CAP67 strain that was reported to lack GXM (50). Although we cannot rule out a cross-reactivity of these antibodies with GalXM, it is possible that these antibodies react with small amounts of GXM or GXM-like polysaccharides that are present in these purified GalXM fractions. This will be discussed further in Discussion.
FIG. 2.
Reactivity of polyclonal antisera and monoclonal antibodies with GXM and GalXM. Reactivity of the polyclonal antisera anti-H99 and anti-GalXM (A) and monoclonal antibodies 18B7 and 13F1 (B) against purified GXM and GalXM was tested in dot blots. Purified polysaccharides (4, 2, or 1 μg for anti-H99 and anti-GalXM and 1 μg or 0.01 μg for monoclonal antibodies 18B7 and 13F1) were spotted onto nitrocellulose membranes and incubated with the indicated antisera or monoclonal antibodies. α, anti-.
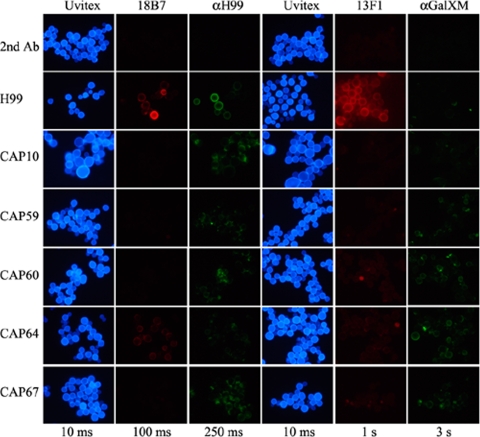
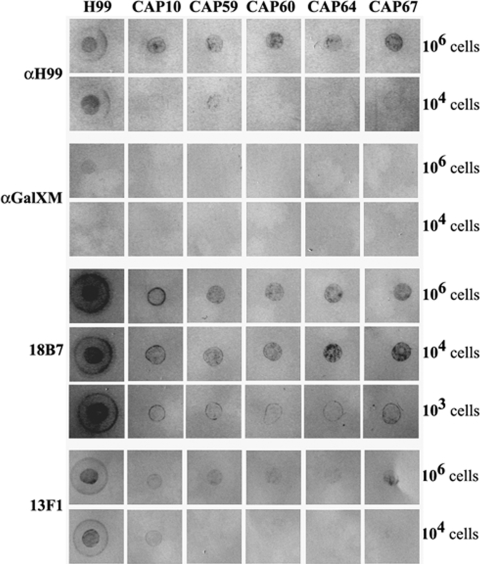
Reactivity of the polyclonal and monoclonal antibodies in cells was assessed by immunofluorescence. Yeast cells were grown in GSU medium, in which capsule production is induced due to iron limitation (16). As expected, wild-type cells, containing both GXM and GalXM in their capsule, reacted with the anti-H99 and anti-GalXM antisera as well as the monoclonal antibodies 18B7 and 13F1 (Fig. 3). Reactivity with GalXM antiserum was rather low, probably due to the small amount of this polysaccharide in the capsule. A similar weak reaction was observed at the cell surfaces of the CAP59, CAP60, CAP64, and CAP67 mutants (Fig. 3), and an exposure time of 3 s was required. Reactivity of the anti-GalXM antiserum with the CAP10 mutant was clearly lower. The CAP mutants also reacted weakly with the anti-H99 antiserum, and a preference for binding was observed at locations preferentially near budding sites. The 18B7 antibody did not bind to the CAP mutants, except for the CAP64 mutant, which showed moderate binding of the antibody (Fig. 3). The 13F1 antibody displayed moderate binding to the CAP60, CAP64, and CAP67 mutants but hardly any or no binding to the cell surfaces of the CAP59 and CAP10 mutants. Dot blot analysis on these cells showed similar results (Fig. 4), except for the 18B7 antibody, where clear staining of the CAP mutant cells was observed when 106 and 104 cells were used. Immunodetection with 103 cells and the 18B7 antibody (Fig. 4) was more in agreement with the results depicted in Fig. 3, and the reaction with the CAP67 mutant was more pronounced than with the other CAP mutants. For the anti-GalXM antiserum, hardly any staining of CAP mutant cells was detected by dot blot analysis (Fig. 4), which is in agreement with the very weak signal seen in Fig. 3. These results indicate that GalXM and/or GXM or GXM-like polysaccharides are present at the surfaces of the CAP mutant cells.
FIG. 3.
Immunofluorescence of cells from the C. neoformans wild type (H99) and CAP mutants. Cells were grown for 5 days in GSU medium and subsequently incubated with Uvitex (cell wall staining), monoclonal antibodies 18B7 and 13F1, and polyclonal antisera anti-H99 and anti-GalXM, as described in Materials and Methods. The top row indicated by “2nd Ab” is the CAP59 mutant incubated only with the second antibody to assess the background signal. Exposure times for each column are indicated. α, anti-.
FIG. 4.
Dot blot analysis of whole cells from the C. neoformans wild type (H99) and CAP mutants grown for 5 days in GSU medium. Rows indicate reactions of different numbers of cells with the 18B7, anti-H99, 13F1, and anti-GalXM antibodies. α, anti-.
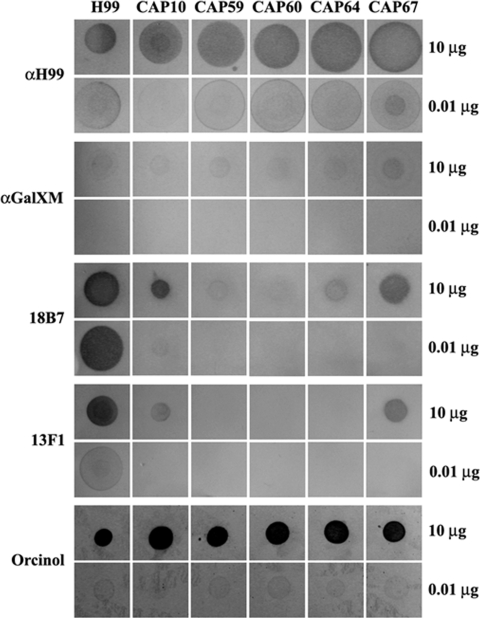
Dot blot analysis was performed to investigate whether GalXM and/or GXM-like polysaccharides are secreted into the culture medium of the CAP mutants. Immunoreactive material was detected in the growth medium of both the wild type and the CAP mutants with the anti-H99, 18B7, and anti-GalXM antibodies (Fig. 5). With the antibody 13F1 hardly any, if any, immunoreactive material was detected in the growth medium of CAP59, CAP60, and CAP64 mutants, but small amounts were detected with the CAP10 and CAP67 mutants (Fig. 5). Taken together, the results indicate that CAP mutants do produce capsular polysaccharides and that a substantial amount of these polysaccharides is secreted into growth medium. Interestingly, these polysaccharides appear to be GalXM as well as GXM-like, based on the reactivity of the GXM- and GalXM-specific antisera.
FIG. 5.
Dot blot analysis of isolated polysaccharides from the C. neoformans wild type (H99) and CAP mutants grown for 5 days in GSU medium. Rows indicate reactions of different amounts of polysaccharides with the 18B7, anti-H99, 13F1, and anti-GalXM antibodies. Orcinol staining of polysaccharides spotted onto TLC plates was used to demonstrate that equal amounts of GXM and GalXM were used in the dot blots. α, anti-.
Composition analysis of polysaccharide released in the growth media.
Monosaccharide analyses were performed to obtain more information on the chemical composition of the polysaccharides secreted by the CAP mutants. Approximately 10 times more polysaccharides with molecular masses of ≥30 kDa were extracted from the medium of the wild-type strain than were extracted from the media of the CAP mutants. The isolated polysaccharides were subjected to methanolysis and analyzed by GLC and GLC MS. Molar ratios of purified GXM agreed with expected ratios from the literature (Table 1; Fig. 1) (14). Monosaccharides of purified GalXM, however, also contained glucose and glucuronic acid, but mannose levels were lower than predicted from the literature (Table 1) (50). The presence of glucose should be considered with care, as glucose is frequently found as a contaminant due to isolation protocols (e.g., purification on DEAE Sephadex). Alternatively or in addition, glucans might be copurified due to association with GalXM, as has been described previously (29). Molar ratios of the monosaccharides contained in the polysaccharide fraction secreted by the wild-type strain H99 agreed with predicted ratios (Table 1) (14, 50), assuming that both GalXM and GXM are secreted. The molar ratios of the monosaccharides present in the polysaccharide fraction of the CAP mutants agreed with the presence of GalXM but not with that of GXM. Only the polysaccharide fraction of the CAP60 mutant contained more mannose, as would be predicted from the presence of GalXM (Table 1). Interestingly, all CAP mutants secreted polysaccharide material containing glucuronic acid, which is normally present in GXM.
TABLE 1.
Molar ratios of monosaccharides in purified fractionated polysaccharidesa
| Fractionated polysaccharide | Molar ratios of indicated monosaccharides in fractionated polysaccharides
|
||||
|---|---|---|---|---|---|
| Mannose | Galactose | Xylose | Glucuronic acid | Glucose | |
| Wild-type H99 (ser. A) | 30.9 ± 7.3 | 6.0 | 18.6 ± 4.2 | 11.6 ± 0.6 | TR |
| CAP10 (ser. A) | 3.4 ± 0.2 | 6.0 | 2.4 ± 0.1 | 1.4 ± 0.4 | TR |
| CAP59 (ser. A) | 4.7 ± 0.2 | 6.0 | 2.3 ± 0.1 | 1.1 ± 0.1 | TR |
| CAP60 (ser. A) | 5.8 ± 0.1 | 6.0 | 2.4 ± 0.1 | 1.2 ± 0.1 | TR |
| CAP64 (ser. A) | 4.6 ± 0.4 | 6.0 | 2.3 ± 0.1 | 1.3 ± 0.4 | TR |
| CAP67 (ser. D) | 4.7 ± 0.1 | 6.0 | 2.4 ± 0.1 | 0.8 ± 0.6 | TR |
| Purified GXM | 3.0 | 2.0 | 1.0 | 0.2 | |
| Purified GalXM | 2.1 | 6.0 | 1.5 | 2.6 | 13.5 |
| GSU (medium) | TR | TR | TR | TR | |
Polysaccharides were purified from GSU culture medium in which different C. neoformans strains were grown for 5 days. Expected ratios of monosaccharides were derived from structures of GXM and GalXM, as depicted in Fig. 1, and galactose was set at six as a reference for all polysaccharide fractions analyzed, except purified GXM, for which mannose was set at three. Carbohydrate content ranged between 400 and 4,000 μg per fraction, except for GSU growth medium, which was too low to be accurately determined. The purified GXM and GalXM fractions were obtained via different purification protocols (15, 20) and enable a comparison of monosaccharide compositions between polysaccharide fractions purified via different methods. ser., serotype; TR, trace.
The polysaccharide fractions of wild-type H99 and the CAP10 and CAP67 mutants were separated by anion-exchange chromatography (Table 2). Based on the charge, GXM is expected to bind to the column, whereas GalXM will not. Small amounts of wild-type polysaccharide did not bind to the column (flowthrough). This fraction contained monosaccharides present in a molar ratio that unexpectedly fitted with GXM but not with GalXM. The majority of the wild-type polysaccharides did bind to the anion-exchange column, and the monosaccharide composition indicated the presence of both GXM and, surprisingly, GalXM. The vast majority of the polysaccharides from CAP67 and CAP10 mutants also bound to the anion-exchange column, which is, for the CAP67 data, in agreement with previous reports (16, 29, 49, 50). In a second, independent polysaccharide isolation from CAP67 mutant culture medium, the molar ratios of the monosaccharides that bound to the anion-exchange column were 3.8:6.0:2.4:1.0 for mannose/galactose/xylose/glucuronic acid, respectively, indicating that glucuronic acid was present in more than trace amounts. Taken together, these data indicate that GalXM from the CAP10 and CAP67 mutants contains glucuronic acid. The small amount of polysaccharides that were detected in the flowthrough hardly contained or did not contain glucuronic acid but consisted of mannose and xylose and therefore could represent GXM-like polymers.
TABLE 2.
Molar ratios of monosaccharides in fractionated polysaccharides by anion-exchange chromatographya
| Fractionated polysaccharides | Molar ratios of indicated monosaccharides in fractionated polysaccharides by anion-exchange chromatography
|
||||
|---|---|---|---|---|---|
| Mannose | Galactose | Xylose | Glucuronic acid | Glucose | |
| Wild-type H99 (ser. A) | |||||
| With flowthrough | 3.0 | 2.0 | 0.4 | TR | |
| With 100 mM NaCl | 3.0 | 2.0 | 1.6 | 0.4 | |
| With 250 mM NaCl | 3.0 | 0.3 | 2.0 | 1.1 | |
| CAP10 (ser. A) | |||||
| With flowthrough | 3.0 | 3.5 | TR | 0.6 | |
| With 100 mM NaCl | 3.3 | 6.0 | 2.3 | 1.6 | |
| With 250 mM NaCl | 3.2 | 6.0 | 2.5 | 0.7 | |
| CAP67 (ser. D) | |||||
| With flowthrough | TR | TR | TR | TR | |
| With 100 mM NaCl | 4.0 | 6.0 | 2.4 | TR | TR |
| With 250 mM NaCl | 4.3 | 6.0 | 2.2 | TR | TR |
Polysaccharides were purified from GSU culture medium of different C. neoformans strains and loaded onto a Q-Sepharose anion-exchange column. Unbound material was collected in the flowthrough fraction. Bound polysaccharides were eluted from the column using washing steps with increasing amounts of NaCl (concentrations used are indicated). ser., serotype; TR, trace.
NMR analysis of polysaccharide released in growth media.
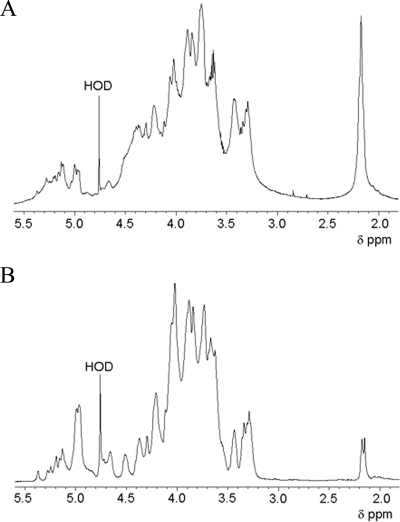
One-dimensional 1H NMR showed that the spectrum of the polysaccharides from the wild-type H99 medium (Fig. 6A) is quite different from those of the CAP mutants (Fig. 6B). A high degree of O acetylation (intense signal at δ, 2.175; shoulder at δ, 2.152) is observed for the wild-type H99 polysaccharides. However, the CAP mutant polysaccharides also showed some degree of O acetylation. Comparable low intensities of the O-acetyl signals at δ of 2.177 and 2.153 were observed for polysaccharides purified from the CAP67, CAP59, CAP60, and CAP64 mutants. In the case of the polysaccharides derived from the CAP10 mutant, the signal at δ of 2.153 occurred as a shoulder of the low-intensity signal at δ of 2.177. Spectra of the wild-type and the CAP mutants were also different with respect to the anomeric peak pattern (δ, 4.5 to 5.5). Notably, those of the CAP mutants were highly similar, indicating that these mutants produce polysaccharides with similar structures. Among the five mutants, only the anomeric peak pattern of the CAP10 mutant polysaccharides differed slightly. The anomeric region of the polysaccharides of the CAP mutants showed similarities with the 1H NMR spectrum of GalXM from the CAP67 mutant (50), indicating that majority of the polysaccharides produced by the CAP mutants represent GalXM.
FIG. 6.
1H NMR spectra (500 MHz) of wild-type (A) and CAP59 mutant (B) polysaccharides isolated from the medium. The spectrum of the CAP59 mutant is representative for all CAP mutants. Differences between the wild type and the CAP mutants can be seen in the deviating anomeric peak pattern (δ, 4.5 to 5.5). HOD, monodeuterated water.
DISCUSSION
Mutants of C. neoformans with deletions in the CAP10, CAP59, CAP60, and CAP64 genes have an acapsular phenotype, and this phenotype is generally believed to be due to the inability to produce GXM, the major component of the capsule polysaccharides. Interestingly, it is shown here that the CAP mutants do produce GalXM and GXM-like molecules. These molecules are present at the cell surfaces of the mutants but are also secreted into the culture medium. It should be noted that the amounts of polysaccharides that are produced by the mutant are 5 to 10 times smaller, which could explain their acapsular phenotype.
Wild-type and CAP mutant cells were grown under capsule-inducing conditions. Wild-type cells reacted with antibodies that recognize GalXM and GXM. Reactivity with the GalXM antiserum was rather low, possibly explained by the smaller amount of this polysaccharide in the capsule. Interestingly, the CAP59, CAP60, CAP64, and CAP67 mutants gave a similar reaction at the cell surface, indicating that GalXM is present with an amount not very different from that in the wild type. The reaction of the CAP10 mutant was clearly lower, which suggests a smaller amount of the polysaccharide at the cell surface of this mutant. The alternative explanation, the presence of an aberrant GalXM molecule, is not very likely, since the GalXM structures of all mutants seem to be identical (see below). Antisera reacting with GXM generally reacted less strongly with the cell surfaces of the CAP mutants than with that of the wild type. This indicates that this polysaccharide is less abundant in these mutant strains. Notably, the GXM antisera reacted differently with the CAP mutants. For instance, the 13F1 monoclonal antibody bound moderately to the cell surfaces of the CAP60, -64, and -67 mutants but not with those of the CAP59 and CAP10 mutants. This may indicate that the latter mutants have a smaller amount of GXM-like polysaccharides at their surfaces or produce an aberrant structure.
Polysaccharides were purified from the medium of wild-type and CAP mutants using a simple purification procedure. The polymer fraction with a molecular mass exceeding 30 kDa was treated with DNase and proteinase, followed with a trichloroacetic acid precipitation and dialyses. The amounts of polysaccharides produced by the wild type and the acapsular mutants agree with the previously reported ratio of GXM to GalXM of about 10:1 (14, 50), based on the observations described in this study that the acapsular mutants produce no GXM but GalXM plus only small amounts of GXM-like polymers. The presence of these GXM-like polymers can explain the reactivity of the monoclonal antibodies 18B7 and 13F1 with GalXM fractions extracted from the medium of CAP mutants. The GXM-like polymers are copurified with GalXM, possibly due to interactions between GXM-like polymers and GalXM, in a similar way to what was recently shown for GXM and GalXM (25). Interestingly, unexpected cell wall labeling with monoclonal antibody 13F1 has been described after immunogold labeling of CAP67 cells (23). The authors suggested the possibility that more than one type of GXM molecule is produced by some strains of Cryptococcus neoformans. That suggestion is in accordance with our findings that small amounts of GXM-like polysaccharides are produced and secreted by Cryptococcus neoformans. We detect these GXM-like polysaccharides in the secreted polysaccharide fractions of the CAP mutants. We cannot exclude the possibility that these GXM-like polysaccharides are also produced by the wild-type strain but are difficult to detect due to the presence of large amounts of GXM.
NMR analysis confirmed the presence of GalXM in the medium of the CAP mutants. The spectra of the polysaccharides were similar to that published for the GalXM structure of the CAP67 strain (50). Of interest, glucuronic acid was found in minor but significant amounts in the secreted polysaccharides of the CAP mutants. This sugar is considered to be a constituent of GXM. This prompted us to purify the polysaccharides by anion-exchange chromatography and analysis of the fractions with GLC MS. In contrast to GalXM, GXM is expected to bind to the column. Indeed, most of the GXM secreted by the wild-type strain bound to the column. The fact that a small fraction did not bind to the column can be explained by entrapment of GXM in vesicles composed of neutral lipids (47). Surprisingly, all GalXM was found to bind to the column, indicating the presence of charged residues or an interaction of the GalXM with GXM. Notably, GalXM of the CAP67 and CAP10 mutants also bound to the column. The monosaccharide molar ratios indicated that GalXM secreted by the mutant strains contains glucuronic acid. This finding is, in fact, in agreement with previously reported data (16, 49). For instance, it has been shown in the past that GalXM in the medium of the CAP67 mutant binds to an anion-exchange column (14, 50). The structure of GalXM has been determined from a purified subfraction. This, together with the technical difficulties of detecting glucuronic acid, might explain why the proposed structure of GalXM lacked glucuronic acid. In this study, we demonstrate that glucuronic acid is present in the GalXM structure of CAP67 and the four other CAP mutants and in the GalXM fraction that was purified from CAP67, according to the protocol of Cherniak et al. (data in Table 1) (15, 20). To further strengthen the finding that glucuronic acid is part of GalXM, we excluded the presence of hyaluronic acid. This polymer that contains glucuronic acid has recently been reported in the capsule of C. neoformans (8, 32). However, we have been unable to detect this polymer in the polysaccharide fraction (J. Grijpstra and H. de Cock, unpublished data). It is concluded that part of the repeating units of GalXM of the CAP mutants, but possibly also of the wild type, is glucuronylated.
A total of 1 to 5% of the polysaccharides in the medium of CAP67 and CAP10 mutants did not bind to the anion-exchange column. This fraction could not be related to GalXM. It consisted of mannose and xylose, with traces of glucose and, if present at all, of glucuronic acid. The molar ratios of monosaccharides in this fraction agree with a GXM-like structure lacking most, if not all, glucuronic acid. Secretion of the GXM-like polymers agrees with the fact that the polysaccharides in the medium reacted with antisera against GXM. 1H NMR spectra indicated the presence of O-acetylated polymers, although the O acetylation of them was found to a lesser extent than that of the wild type. We have used PI staining to determine if the increased cell death and lysis could be responsible for the GXM-like polysaccharides in the culture medium of the wild type and CAP10 and CAP59 mutants. However, staining of cells grown for 1 or 5 days with PI (47) indicated virtually 100% viable cells. Only prolonged growth at 30°C revealed a small increase in dead cells (at 10 days, approximately 10%; at 21 days, up 20%). The extremely small amount of dead cells at day 5 does not seem to correlate with the amount of the GXM-like polysaccharides that is detected in the culture medium of the cells. We therefore propose that GXM-like polysaccharides are indeed produced and secreted by the CAP mutants. We would like to point out that structural analysis of polysaccharides of Cryptococcus have been reported to be performed on polysaccharides purified from culture grown for a few day up to 2 weeks (16, 18, 39). The possibility exists that Cryptococcus produces small amounts of different polysaccharides to vary its surface structure in order to evade the immune system.
The results that have been described indicate that deletion of the CAP genes affects especially the production of GXM rather than that of GalXM. This would agree with the 10-fold-smaller amount of polysaccharide found in the medium of the CAP mutants, since GXM is approximately 10-fold more abundant in the wild type than GalXM. The abundance of this polysaccharide also explains why all CAP mutants are affected in the synthesis of GXM. Chang and Kwon-Chung (9) selected for mutants lacking a clearly visible capsule. Mutants lacking only GalXM would not be selected in this screening method because GalXM does not affect the appearance of the extracellular capsule matrix (41). It has been reported that Cap59/Cap60 and Cap10 share homology with mannosyl and xylosyl transferases, respectively (34, 48). Such transferases could also be involved in the formation of GalXM. Our data suggest that both of the capsule polysaccharides have their own machinery for synthesis. However, there may be some overlap, which may explain the presence of glucuronic acid in (part of) the GalXM molecules.
ADDENDUM
Recently, the extracellular polysaccharides from a CAP67 mutant have been reanalyzed (28), and the researchers found that one of the residues that is 3 linked to the side chain galactose is not β-d-xylose, but is β-d-glucuronic acid, confirming our results.
Acknowledgments
We are indebted to Guilhem Janbon (Institute Pasteur, Paris, France) for the C. neoformans strains, Arturo Casadevall (Albert Einstein College of Medicine, NY) for the 18B7 and 13F1 monoclonal antibodies, Christophe Blanchetot (Utrecht University, Utrecht) for the S. cerevisiae strain, Slavomír Bystrický (Institute of Chemistry, Slovak Academy of Sciences) for the anti-GalXM antiserum, Frank Coenjaerts (Utrecht Medical Center, Utrecht) for the anti-H99 antiserum, purified GXM, purified GalXM, and the CAP67 strain, Stefan Oscarson for the tetrasaccharide used in GLC MS analysis, and S. P. Jongen (Department of Bio-Organic Chemistry, Utrecht University) for recording the 1H NMR spectra. We thank Menno Lammers for assistance in PI experiments.
Footnotes
Published ahead of print on 19 June 2009.
REFERENCES
- 1.Beverley, S. M., K. L. Owens, M. Showalter, C. L. Griffith, T. L. Doering, V. C. Jones, and M. R. McNeil. 2005. Eukaryotic UDP-galactopyranose mutase (GLF gene) in microbial and metazoal pathogens. Eukaryot. Cell 41147-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharjee, A. K., J. E. Bennett, and C. P. Glaudemans. 1984. Capsular polysaccharides of Cryptococcus neoformans. Rev. Infect. Dis. 6619-624. [DOI] [PubMed] [Google Scholar]
- 3.Bose, I., A. J. Reese, J. J. Ory, G. Janbon, and T. L. Doering. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretaudeau, K., O. Eloy, A. Richer, F. Bruneel, D. Scott-Algara, O. Lortholary, and F. Pico. 2006. Cryptococcal meningo-encephalitis in an apparently immunocompetent patient. Rev. Neurol. (Paris) 162233-237. (In French.) [DOI] [PubMed] [Google Scholar]
- 5.Buchanan, K. L., and J. W. Murphy. 1998. What makes Cryptococcus neoformans a pathogen? Emerg. Infect. Dis. 471-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bystricky, S., E. Paulovicova, and E. Machova. 2004. Synthesis and immunogenicity of polysaccharide-protein conjugate composed of galactoglucoxylomannan of Cryptococcus laurentii. FEMS Microbiol. Lett. 235311-314. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Y. C., R. Cherniak, T. R. Kozel, D. L. Granger, L. C. Morris, L. C. Weinhold, and K. J. Kwon-Chung. 1997. Structure and biological activities of acapsular Cryptococcus neoformans 602 complemented with the CAP64 gene. Infect. Immun. 651584-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. C., A. Jong, S. Huang, P. Zerfas, and K. J. Kwon-Chung. 2006. CPS1, a homolog of the Streptococcus pneumoniae type 3 polysaccharide synthase gene, is important for the pathobiology of Cryptococcus neoformans. Infect. Immun. 743930-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 144912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. C., and K. J. Kwon-Chung. 1998. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 662230-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, Y. C., and K. J. Kwon-Chung. 1999. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 1815636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 1996. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 641977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, Y. C., B. L. Wickes, and K. J. Kwon-Chung. 1995. Further analysis of the CAP59 locus of Cryptococcus neoformans: structure defined by forced expression and description of a new ribosomal protein-encoding gene. Gene 167179-183. [DOI] [PubMed] [Google Scholar]
- 14.Cherniak, R., R. G. Jones, and E. Reiss. 1988. Structure determination of Cryptococcus neoformans serotype A-variant glucuronoxylomannan by 13C-n.m.r. spectroscopy. Carbohydr. Res. 172113-138. [DOI] [PubMed] [Google Scholar]
- 15.Cherniak, R., L. C. Morris, B. C. Anderson, and S. A. Meyer. 1991. Facilitated isolation, purification, and analysis of glucuronoxylomannan of Cryptococcus neoformans. Infect. Immun. 5959-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherniak, R., E. Reiss, and S. H. Turner. 1982. A galactoxylomannan antigen of Cryptococcus neoformans serotype A. Carbohydr. Res. 103239-250. [Google Scholar]
- 17.Cherniak, R., and J. B. Sundstrom. 1994. Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect. Immun. 621507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherniak, R., H. Valafar, L. C. Morris, and F. Valafar. 1998. Cryptococcus neoformans chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronoxylomannans with a computer-simulated artificial neural network. Clin. Diagn. Lab Immunol. 5146-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chute, H. L., F. L. Davis, and D. D. Payne. 1965. Cryptococcosis, a disease of man and animals. J. Maine Med. Assoc. 56228-229. [PubMed] [Google Scholar]
- 20.Coenjaerts, F. E., A. M. Walenkamp, P. N. Mwinzi, J. Scharringa, H. A. Dekker, J. A. van Strijp, R. Cherniak, and A. I. Hoepelman. 2001. Potent inhibition of neutrophil migration by cryptococcal mannoprotein-4-induced desensitization. J. Immunol. 1673988-3995. [DOI] [PubMed] [Google Scholar]
- 21.Doering, T. L. 2000. How does Cryptococcus get its coat? Trends Microbiol. 8547-553. [DOI] [PubMed] [Google Scholar]
- 22.Ellerbroek, P. M., D. J. Lefeber, R. van Veghel, J. Scharringa, E. Brouwer, G. J. Gerwig, G. Janbon, A. I. Hoepelman, and F. E. Coenjaerts. 2004. O-acetylation of cryptococcal capsular glucuronoxylomannan is essential for interference with neutrophil migration. J. Immunol. 1737513-7520. [DOI] [PubMed] [Google Scholar]
- 23.Feldmesser, M., Y. Kress, and A. Casadevall. 2001. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology 1472355-2365. [DOI] [PubMed] [Google Scholar]
- 24.Franzot, S. P., I. F. Salkin, and A. Casadevall. 1999. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J. Clin. Microbiol. 37838-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frases, S., L. Nimrichter, N. B. Viana, A. Nakouzi, and A. Casadevall. 2008. Cryptococcus neoformans capsular polysaccharide and exopolysaccharide fractions manifest physical, chemical, and antigenic differences. Eukaryot. Cell 7319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Rivera, J., Y. C. Chang, K. J. Kwon-Chung, and A. Casadevall. 2004. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 3385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton, A. J., and J. Goodley. 1996. Virulence factors of Cryptococcus neoformans. Curr. Top. Med. Mycol. 719-42. [PubMed] [Google Scholar]
- 28.Heiss, C., J. S. Klutts, Z. Wang, T. L. Doering, and P. Azadi. 2009. The structure of Cryptococcus neoformans galactoxylomannan contains beta-D-glucuronic acid. Carbohydr. Res. 344915-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James, P. G., and R. Cherniak. 1992. Galactoxylomannans of Cryptococcus neoformans. Infect. Immun. 601084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janbon, G. 2004. Cryptococcus neoformans capsule biosynthesis and regulation. FEMS Yeast Res. 4765-771. [DOI] [PubMed] [Google Scholar]
- 31.Janbon, G., U. Himmelreich, F. Moyrand, L. Improvisi, and F. Dromer. 2001. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42453-467. [DOI] [PubMed] [Google Scholar]
- 32.Jong, A., C. H. Wu, H. M. Chen, F. Luo, K. J. Kwon-Chung, Y. C. Chang, C. W. Lamunyon, A. Plaas, and S. H. Huang. 2007. Identification and characterization of CPS1 as a hyaluronic acid synthase contributing to the pathogenesis of Cryptococcus neoformans infection. Eukaryot. Cell 61486-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klutts, J. S., and T. L. Doering. 2008. Cryptococcal xylosyltransferase 1 (Cxt1p) from Cryptococcus neoformans plays a direct role in the synthesis of capsule polysaccharides. J. Biol. Chem. 28314327-14334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klutts, J. S., S. B. Levery, and T. L. Doering. 2007. A beta-1,2 xylosyltranferase from Cryptococcus neoformans defines a new family of glycosyltransferases. J. Biol. Chem. 28217890-17899. [DOI] [PubMed] [Google Scholar]
- 35.Kozel, T. R. 1995. Virulence factors of Cryptococcus neoformans. Trends Microbiol. 3295-299. [DOI] [PubMed] [Google Scholar]
- 36.Levitz, S. M. 1991. The ecology of Cryptococcus neoformans and the epidemiology of cryptococcosis. Rev. Infect. Dis. 131163-1169. [DOI] [PubMed] [Google Scholar]
- 37.Levitz, S. M., D. J. DiBenedetto, and R. D. Diamond. 1987. A rapid fluorescent assay to distinguish attached from phagocytized yeast particles. J. Immunol. Methods 10137-42. [DOI] [PubMed] [Google Scholar]
- 38.Lui, G., N. Lee, M. Ip, K. W. Choi, Y. K. Tso, E. Lam, S. Chau, R. Lai, and C. S. Cockram. 2006. Cryptococcosis in apparently immunocompetent patients. QJM 99143-151. [DOI] [PubMed] [Google Scholar]
- 39.McFadden, D. C., B. C. Fries, F. Wang, and A. Casadevall. 2007. Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans. Eukaryot. Cell 61464-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moyrand, F., Y. C. Chang, U. Himmelreich, K. J. Kwon-Chung, and G. Janbon. 2004. Cas3p belongs to a seven-member family of capsule structure designer proteins. Eukaryot. Cell 31513-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moyrand, F., T. Fontaine, and G. Janbon. 2007. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol. Microbiol. 64771-781. [DOI] [PubMed] [Google Scholar]
- 42.Moyrand, F., and G. Janbon. 2004. UGD1, encoding the Cryptococcus neoformans UDP-glucose dehydrogenase, is essential for growth at 37°C and for capsule biosynthesis. Eukaryot. Cell 31601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee, J., A. Casadevall, and M. D. Scharff. 1993. Molecular characterization of the humoral responses to Cryptococcus neoformans infection and glucuronoxylomannan-tetanus toxoid conjugate immunization. J. Exp. Med. 1771105-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy, J. W. 1999. Immunological down-regulation of host defenses in fungal infections. Mycoses 42(Suppl. 2)37-43. [PubMed] [Google Scholar]
- 45.Perfect, J. R., B. Wong, Y. C. Chang, K. J. Kwon-Chung, and P. R. Williamson. 1998. Cryptococcus neoformans: virulence and host defences. Med. Mycol. 36(Suppl. 1)79-86. [PubMed] [Google Scholar]
- 46.Reiss, E., M. Huppert, and R. Cherniak. 1985. Characterization of protein and mannan polysaccharide antigens of yeasts, moulds, and actinomycetes. Curr. Top. Med. Mycol. 1172-207. [DOI] [PubMed] [Google Scholar]
- 47.Rodrigues, M. L., L. Nimrichter, D. L. Oliveira, S. Frases, K. Miranda, O. Zaragoza, M. Alvarez, A. Nakouzi, M. Feldmesser, and A. Casadevall. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 648-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sommer, U., H. Liu, and T. L. Doering. 2003. An alpha-1,3-mannosyltransferase of Cryptococcus neoformans. J. Biol. Chem. 27847724-47730. [DOI] [PubMed] [Google Scholar]
- 49.Turner, S. H., R. Cherniak, and E. Reiss. 1984. Fractionation and characterization of galactoxylomannan from Cryptococcus neoformans. Carbohydr. Res. 125343-349. [DOI] [PubMed] [Google Scholar]
- 50.Vaishnav, V. V., B. E. Bacon, M. O'Neill, and R. Cherniak. 1998. Structural characterization of the galactoxylomannan of Cryptococcus neoformans Cap67. Carbohydr. Res. 306315-330. [DOI] [PubMed] [Google Scholar]
- 51.Yoneda, A., and T. L. Doering. 2006. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol. Biol. Cell 175131-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zebedee, S. L., R. K. Koduri, J. Mukherjee, S. Mukherjee, S. Lee, D. F. Sauer, M. D. Scharff, and A. Casadevall. 1994. Mouse-human immunoglobulin G1 chimeric antibodies with activities against Cryptococcus neoformans. Antimicrob. Agents Chemother. 381507-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]