FIG. 6.
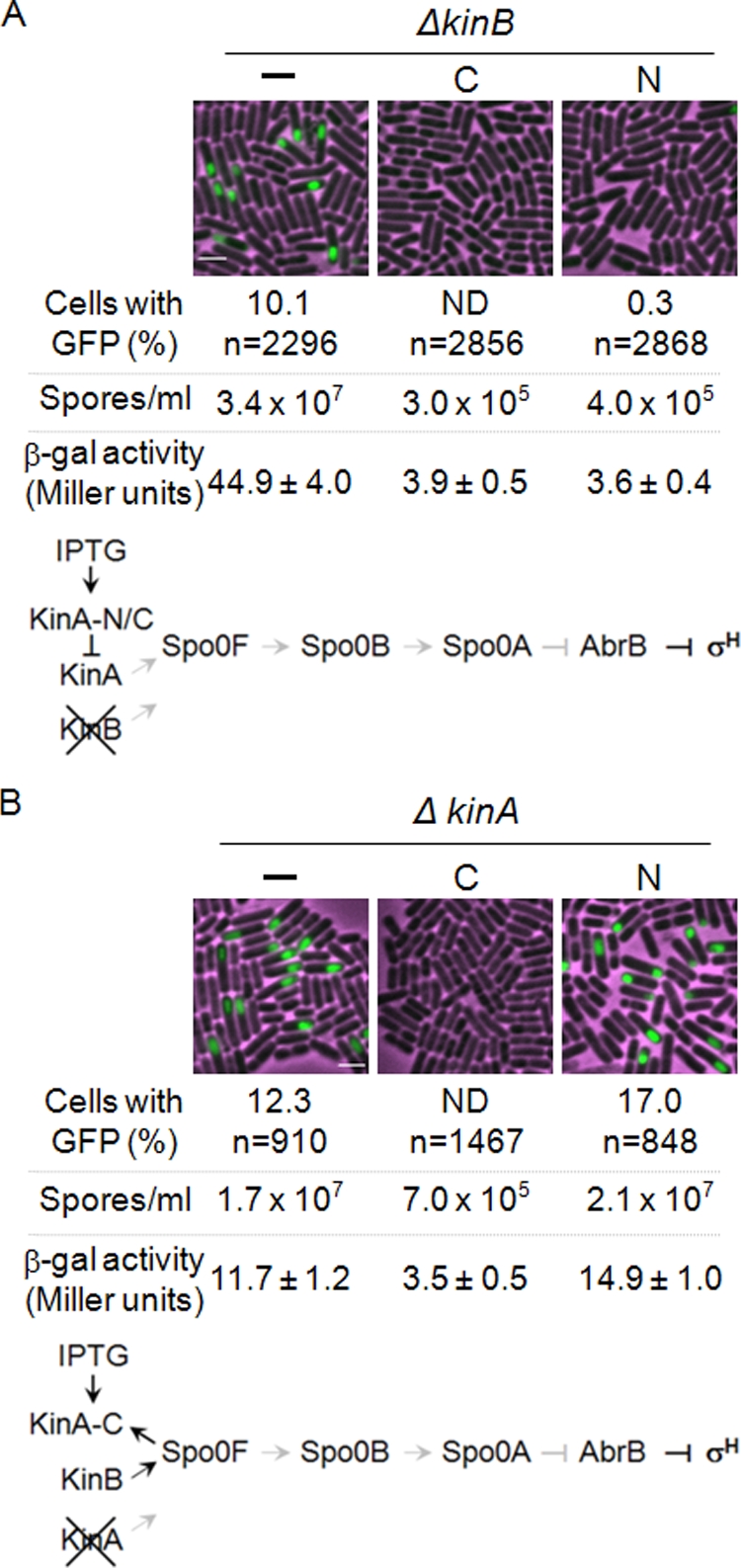
Effects of overexpression of the N- and C-terminal domains of KinA on sporulation. (A) Dominant-negative assay. The gene encoding the truncated KinAN or KinAC protein was fused to Phyper-spank and introduced at the amyE locus into cells containing the kinB-null mutation. The resulting strains have the gene for wild-type KinA at the endogenous locus and the engineered gene for the mutant at the amyE locus in the kinB-null background. Strains with the kinB-null mutation and lacking the IPTG-inducible construct were used as controls (−). C, KinAC; N, KinAN. Transcriptional reporters were constructed in which the genes encoding β-galactosidase (lacZ) and GFP were fused to the promoter region of the spoIIG operon to monitor Spo0A activity and to the promoter region of the spoIIQ operon to monitor forespore-specific σF activity, respectively. Individual reporters were transferred to the thrC locus of each strain and used for the β-galactosidase assay and GFP image analysis, respectively. The percentage of cells with the GFP signal and the total number of cells scored for GFP image analysis (n) are given. Sporulation efficiency was determined as described in Materials and Methods. The accumulation of β-galactosidase from a PspoIIG-lacZ fusion was monitored at hour 3 after suspension as a measure of Spo0A activity. Data for β-galactosidase are averages of three independent results with standard deviations. Strains used were MF3363 (control) (GFP assay), MF3366 (control) (β-galactosidase and sporulation assays), MF3364 (KinAC) (GFP assay), MF3367 (KinAC) (β-galactosidase and sporulation assays), MF3365 (KinAN) (GFP assay), and MF3368 (KinAN) (β-galactosidase and sporulation assays). ND, not detected. The genetic system that allows examination of the dominant-negative effects and the results are diagramed at the bottom. Arrows and T-shaped bars indicate positive and negative actions, respectively. Black and gray symbols represent active and inactive pathways, respectively. (B) Reverse phosphotransfer reaction assay. Each engineered gene construct, as described for panel A, was introduced at the amyE locus into cells containing the kinA-null mutation. The resulting strains have the gene for wild-type KinB at the endogenous locus and the engineered gene for the mutant at the amyE locus in the kinA-null background. Strains with the kinA-null mutation and lacking the IPTG-inducible construct were used as controls. Transcriptional reporters were constructed as described in the legend to panel A. Strains used were MF3386 (control) (GFP assay), MF2678 (control) (β-galactosidase and sporulation assays), MF3387 (KinAC) (GFP assay), MF2831 (KinAC) (β-galactosidase and sporulation assays), MF3388 (KinAN) (GFP assay), and MF3251 (KinAN) (β-galactosidase and sporulation assays). Bars, 2 μm. The genetic system that allows examination of the reverse phosphotransfer reaction and the results are diagramed at the bottom. We note that the sporulation efficiencies and β-galactosidase activities from the PspoIIG-lacZ reporter in the control strain were slightly lower in the presence of 0.5 mM IPTG than in its absence, while the strains shown in Fig. 3 displayed normal levels with or without 10 μM IPTG.
