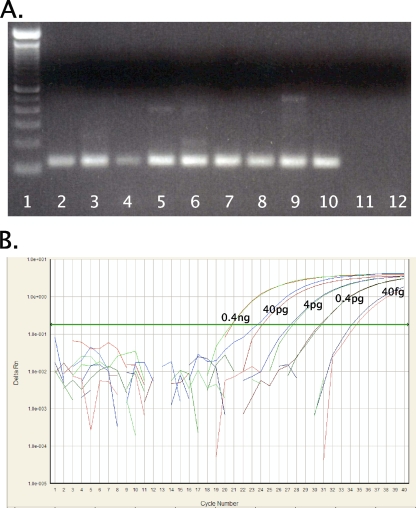
FIG. 1.
RT-PCR for H1 subtype HAs. (A) RT-PCR amplification with the universal H1 HA primer set (set 3). cDNAs from swine, human, and avian origin H1 subtype influenza A viruses were amplified. The final reaction mixture contained 1× PCR Gold buffer, 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphate mixture, 4 U AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA), and 0.6 μM primers. Thermocycling was performed in a DNAEngine (Bio-Rad, Hercules, CA) under the following cycling conditions: 10 min at 95°C, followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. PCR products were electrophoresed on a 2% Tris-acetate-EDTA agarose gel, stained with ethidium bromide, and photographed under UV transillumination. Lanes (left to right): 1, 100-bp DNA molecular weight ladder; 2, A/swine/Ohio/23/1935(H1N1), 3, A/swine/Jamesburg/1942 (H1N1), 4, A/swine/Wisconsin/1/1967 (H1N1); 5, A/Maryland/NIH-37/2009 (H1N1); 6, A/Maryland/NIH-39/2009 (H1N1); 7, A/mallard/Ohio/171/1990 (H1N1); 8, A/green-winged teal/Ohio/72/1999(H1N1); 9, A/green-winged teal/Ohio/430/1987 (H1N1); 10, A/California/04/2009 (H1N1); 11, A/New York/470/2004 (H3N2); 12, water. (B) Amplification curve for influenza A virus H1 HA real-time RT-PCR assay. Shown are the 10−1 to 10−5 dilutions of A/California/04/2009 (H1N1) cDNA (final concentrations, 0.4 ng to 40 fg viral RNA per reaction mixture) with the FAM (novel swine origin) probe. Reactions were run in duplicate. The final reaction mixture contained 1× ABI master mix (Applied Biosystems), 0.9 μM primers, and 0.25 μM of each probe under the following cycling conditions: 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.