Figure 6.
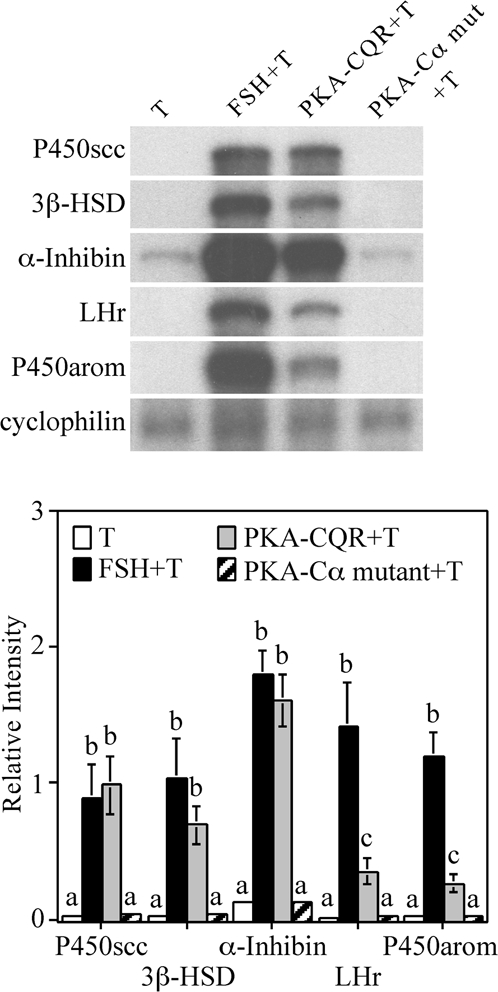
RNase Protection Assay of mRNAs Associated with Granulosa Cell Differentiation in Granulosa Cells Infected with PKA-CQR or PKA-Cα Mutant Lentiviral Vectors
Granulosa cells were infected with either the PKA-CQR or PKA-Cα mutant lentiviral vectors (5 MOI) as described in Fig. 1. The next morning, unattached cells and free lentivirus were removed and replaced with M199. Twenty-four hours later, medium was replaced with fresh medium containing 30 ng/ml testosterone with or without FSH (100 ng/ml). After 48 h, total RNA was extracted from monolayers and analyzed for mRNA by RNase protection assay. The housekeeping gene cyclophilin was included as a loading control. Results shown are representative of three separate groups of granulosa cells (top). Densitometry analysis of the RNase protection assay was performed using the NIH Image software (bottom). The signal of each mRNA was normalized to the housekeeping gene cyclophilin (relative intensity). Results show mean ± 1 sem of three groups of granulosa cells. Significant differences were considered when P < 0.05 and are indicated by different letters in the figure.
