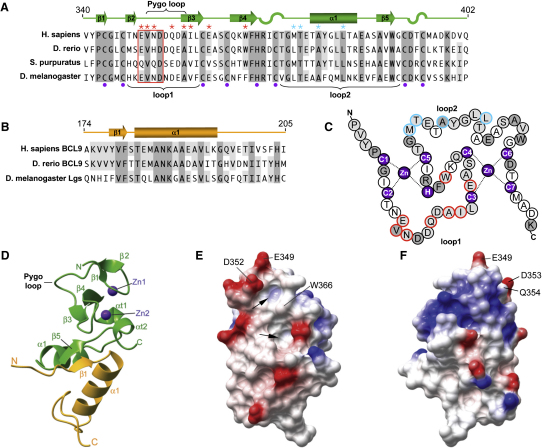
Figure 1.
Sequence Alignments and Structure of the Human PHD-HD1 Complex
(A and B) Alignments of (A) PHD sequences of hPygo1 (Q9Y3Y4), zebrafish Pygo2 (Q1L8T6), sea urchin Pygo (XP_791313), and Drosophila Pygo (Q9V9W8), and (B) HD1 sequences of hBCL9 (O00512), zebrafish BCL9 (Q67FY0), and Drosophila Lgs (Q961D9). Dark gray, invariant residues; light gray, semiconserved residues. Marked above sequences are secondary structure elements (β sheets, α helices; α turns are marked by S shapes) and residues involved in HD1 (blue) or H3K4me binding (red) as defined by mutational analysis (Figures 3B and 4E). EVND motif is boxed; indicated are also Zn2+-coordinating residues (purple), Pygo loop, and loop1 and loop2 surfaces (brackets).
(C) Crossbrace ligation of hPygo1 PHD finger, with Zn-coordinating and mutated residues highlighted (colors as in [A]).
(D) Ribbon representation of the hPHD-HD1 complex structure solved at 1.59 Å resolution, with secondary structure elements labeled as in (A) and (B). PHD, green; HD1, orange; Zn2+, purple.
(E and F) Molecular surface representations of hPHD-HD1, colored according to electrostatic potential (red, negative charges; blue, positive charges), with some Pygo loop and EVND residues labeled. (E) View similar to (D), showing the PHD loop1 surface, with two conspicuous cavities (arrows) separated by W366; (F) is rotated 180° with respect to (E).