Abstract
In enteric bacteria, the cellular response to oxidative stress is activated by oxidation of the iron-sulfur clusters in SoxR, which then induces transcription of soxS, turning on a battery of defense genes. Here we demonstrate both in vitro and in cells that activation of SoxR can occur in a DNA-mediated reaction with guanine radicals, an early genomic signal of oxidative stress, serving as the oxidant. SoxR in its reduced form is found to inhibit guanine damage by repairing guanine radicals. Moreover, cells treated with a DNA-binding photooxidant, which generates guanine radicals, promotes the expression of soxS. In vitro, this photooxidant, tethered to DNA 80 bp from the soxS promoter, induces transcription by activating SoxR upon irradiation. Thus, transcription can be activated from a distance through DNA-mediated charge transport. This chemistry offers a general strategy for DNA-mediated signaling of oxidative stress.
Keywords: DNA charge transport, guanine radicals, oxidative stress
SoxR is a 17-kDa transcription factor responsible for the soxRS response to oxidative stress in bacteria (1). In enteric bacteria, the target gene for SoxR is soxS, a transcription factor that promotes the expression of several dozen genes that play various roles in mitigating oxidative stress in cells (2). SoxR, as a sensor of oxidative stress, shares sequence homology with the MerR family of metal-sensing protein dimers. In vivo, SoxR is maintained in its reduced, inactive form, but becomes rapidly activated under oxidative conditions (3). Importantly, each monomer of SoxR contains a [2Fe2S] cluster that is not critical for DNA binding (4); apo-SoxR binds DNA with comparable affinity to the intact iron protein (Kd = 4.5 × 10−10 M), both in its reduced and oxidized forms (5, 6). The cluster and specifically its oxidation state are, however, essential for the transcriptional activity of SoxR (7, 8). The [2Fe2S] clusters in SoxR each exist in either a reduced +1 state or an oxidized +2 state, and their 1-electron oxidation is responsible for turning on the transcriptional activity. Experiments suggest that DNA-bound oxidation is associated with a significant conformational change in the protein triggering transcriptional activation (9–11).
Even though oxidative stress promotes the activation of SoxR, where in the cell and how SoxR oxidation occur are unclear. SoxR has been shown to respond to agents that generate superoxide (O2−·) (3), but the direct oxidant(s) for the protein is not known. Since the midpoint potential of free SoxR is reported to be −290 mV versus NHE, many cellular oxidants would be available to promote its oxidation, even without oxidative stress. However, DNA binding is associated with a shift in the SoxR midpoint potential of ≈+0.5 V, so that, bound to DNA, it is the reduced form of the protein that is favored in the cellular milieu in the absence of oxidative stress (12); on that basis, one might argue that it is the DNA-bound form that serves as the oxidative switch. There is also no evidence that the superoxide anion radical interacts directly with the [2Fe2S] cluster of SoxR, even though many agents that generate O2−· have been implicated in SoxR activation. Indeed the superoxide radical has been reported in at least one case to destroy the [2Fe2S] cluster of a protein (13). Furthermore, SoxR can be activated by agents that are known to promote DNA damage, such as hydrogen peroxide (14), but, with these agents, rather than reacting with DNA directly, SoxR activation has been attributed to changing the redox balance within the cell.
We have considered that a plausible route for protein oxidation might arise in a DNA-mediated reaction (Fig. 1). Under oxidative stress, reactive oxygen species are generated in the cell that, either directly or indirectly as with the superoxide radical (15), promote reactions that oxidatively damage DNA. Indeed, an early signal of oxidative stress within the cell is the formation of guanine radicals; a generated hole, formed in DNA through reaction with oxidative species, can readily migrate and rapidly equilibrate at guanine sites of low oxidation potential, before slow, irreversible oxidation occurs to form 8-oxo-guanine and other base oxidation products (16, 17). One-electron SoxR oxidation by the guanine radical could both activate transcription factors that respond to oxidative stress and directly repair the base radical. DNA-mediated charge transport has been demonstrated over long molecular distances in a reaction that is sensitive to perturbations in the base pair stack as may arise with mismatches, lesions, and protein binding (18–21). The oxidation of DNA-bound proteins from a distance through DNA charge transport has, furthermore, been established for MutY (22), a base excision repair protein in Escherichia coli, and for p53 (23), a mammalian regulator of the cellular response to genotoxic stress. Electrochemical experiments using DNA-modified electrodes show that the [2Fe2S] clusters of SoxR are electronically well coupled to the DNA base stack with a DNA-bound potential of +200 mV versus NHE (12). With this potential, guanine radicals can oxidize DNA-bound SoxR with high driving force (22). Here we describe the transcriptional activation of SoxR from a distance in vitro through DNA-mediated charge transport and in vivo using a 1 electron DNA-binding photooxidant. Long-range DNA charge transport from guanine radicals provides an effective means to signal oxidative stress across the genome and, in so doing, to activate a response.
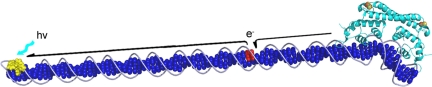
Fig. 1.
Schematized model of transcriptional activation of SoxR from a distance through DNA-mediated charge transport. Here a tethered metal complex (yellow) is used to inject an electron hole into the DNA base pair stack (dark blue) so as to generate a guanine radical (red) (1). DNA-mediated charge transport from SoxR (light blue) (2), bound at its promoter site, to the guanine radical fills the hole and leads to oxidation and activation of SoxR. The SoxR structure shown is based upon the crystal structure of oxidized SoxR bound to DNA (10).
Results
Inhibition of Guanine Damage by SoxR.
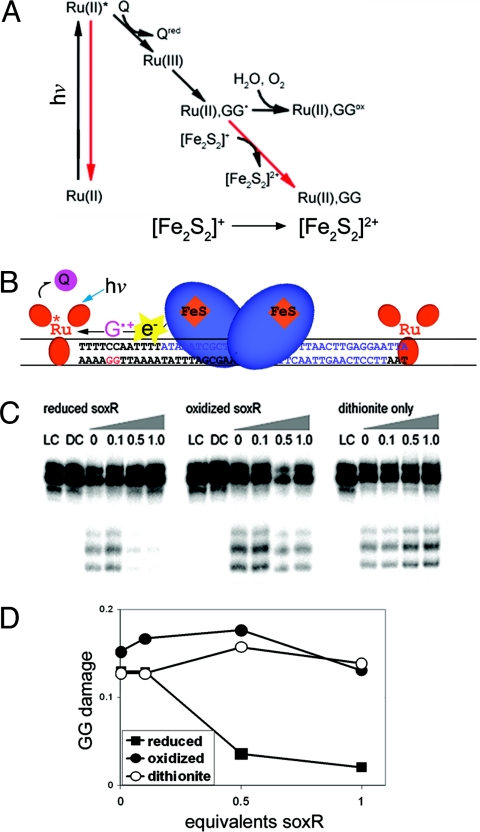
We first examined whether guanine radicals can be reduced by SoxR. Guanine radicals in DNA can be generated efficiently using ruthenium chemistry in a flash-quench scheme (Fig. 2) (24). In this scheme, oxidative quenching of [Ru(phen)(dppz)(bpy′)]2+ (dppz = dipyridophenazine), an avid DNA intercalator, yields a ground state Ru(III) in situ that oxidizes guanine doublets in DNA, generating guanine radicals and consequently a high yield of irreversible guanine damage (21, 25). [Co(NH3)5Cl]2+ is used as a sacrificial excited state quencher, as it precludes back electron transfer, allowing a high yield of damage to be obtained. Reactive oxygen species such as hydroxyl radicals are not produced with this chemistry, and quenching of the excited state prevents the sensitization of singlet oxygen (1O2) (24). Oxidation of a DNA-bound protein can be accomplished by the guanine radicals to intercept irreversible guanine damage. This inhibition of guanine damage has been demonstrated using DNA-bound MutY (22).
Fig. 2.
Inhibition of guanine damage by reduced SoxR. (A) In the Ru flash quench scheme, ground state Ru(II) bound to DNA is irradiated at 442 nm to form excited Ru(II)*. The excited state is quenched by [Co(NH3)5Cl]2+ (Q), resulting in Ru(III) in situ, a ground state oxidant able to abstract an electron from guanine doublets in DNA. If the resulting guanine radical is allowed to react with water, permanent damage products, such as 8-oxo-guanine, form (black arrows). Pathways that prevent formation of damage (red arrows) are recombination processes and/or donation of an electron from the reduced [2Fe2S]+ cluster of SoxR to form [2Fe2S]2+. (B) The experimental system is shown schematically (see Methods). SoxR (blue) is bound to its promoter site on DNA. [Ru(phen)(dppz)(bpy′)]2+ (red), noncovalently intercalated into DNA, is able to oxidize a guanine residue, and the [2Fe2S]+ cluster of SoxR can donate an electron to fill the resulting hole. (C) Denaturing PAGE analysis of guanine oxidation is shown. The DNA used is a 48-mer containing the SoxR binding site. The DNA is 32P-labeled on the 5′-end of the GG containing strand. Concentrations used are 0.5 μM DNA, 5 μM [Ru(phen)(dppz)(bpy′)]2+, and 500 μM [Co(NH3)5Cl]2+. To reduce SoxR, dithionite is added at 10-fold the concentration of SoxR. LC indicates the absence of Ru; DC indicates the absence of light; numbers indicate equivalents of E. coli SoxR:DNA or, for dithionite only, the amount of reductant that would accompany added SoxR. Samples were irradiated for 20 min at 442 nm using a He/Cd laser. After reaction, DNA samples were treated with piperidine to reveal DNA damage as strand breaks; the band above the parent band reflects incomplete reaction with piperidine. The lower bands reflect characteristic oxidative damage at the 5′-GGA-3′ sites. While some decreased damage is evident at very high protein ratios, both with oxidized and reduced protein, consistent with competition between the protein and non-covalent ruthenium for DNA, significant inhibition of damage is evident preferentially for reduced SoxR. D. Quantitation of damage at guanines is shown standardized against the parent band. Uncertainties in the measurement are estimated to be 5%. Guanine damage is greatly attenuated as increasing amounts of reduced SoxR are added to the DNA. This effect is not seen with either oxidized SoxR or dithionite only.
Fig. 2 shows the results of this experiment testing whether reduced SoxR can similarly inhibit guanine damage. DNA damage is generated at guanine doublets, sites in DNA that are most readily oxidized (26), on a DNA duplex using noncovalently bound [Ru(phen)(dppz)(bpy′)]3+ (bpy′ = 4-butyric acid 4′-methyl bipyridine). Oxidative damage is evident only in the presence of DNA-bound Ru, quencher and light, and is absent in samples lacking Ru (light control, LC) or samples containing Ru but not irradiated (dark control, DC). Most interesting is the comparison of damage patterns in the presence of SoxR in different oxidation states. When the DNA is irradiated in the presence of SoxR that has been reduced with sodium dithionite, a large attenuation of the damage at guanine residues is observed, indicating that SoxR is able to reduce the guanine radicals and inhibit irreversible damage. This attenuation increases with increased stochiometric ratios of SoxR to DNA. However, when the DNA is irradiated in the presence of oxidized SoxR, this attenuation in damage is not observed; oxidized SoxR cannot fill the electron hole in the guanine radical. As a control, DNA is irradiated also in the presence of dithionite used to reduce the SoxR; without SoxR, these samples show no attenuation of damage, indicating that it is not the presence of the reductant in solution that is responsible for the attenuation of guanine damage.
These results indicate that the [2Fe2S] clusters in SoxR electronically access the DNA base stack and repair oxidative radical damage at guanine sites in DNA. Since SoxR is a redox sensor involved in mediation of oxidative damage in cells, the ability of SoxR to sense radicals in DNA suggests that guanine damage in DNA could be an initial trigger that allows SoxR to detect and respond to harmful oxidizing conditions in the genome.
Activation of SoxR in Vivo by a DNA Photooxidant.
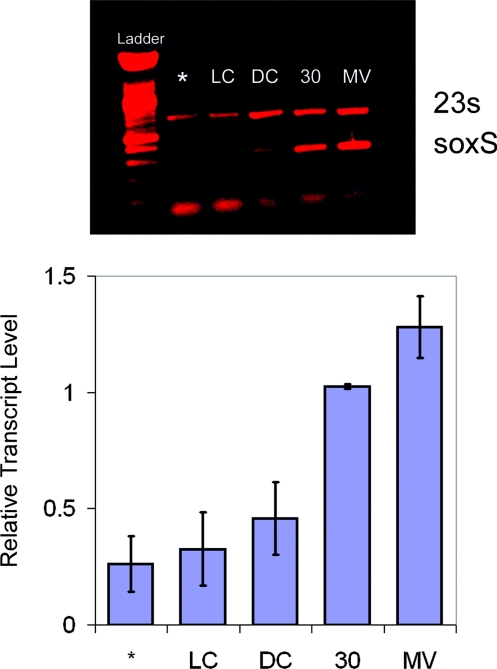
To test whether SoxR can be activated in cells by specifically inducing DNA damage, we examined expression of soxS in wild type Escherichia coli. A single starter culture of E. coli was used to inoculate cultures in media containing or lacking [Rh(phi)2bpy]3+ (phi = 9,10-phenanthrenequinone diimine). This rhodium complex also binds DNA tightly by intercalation, is a potent photooxidant, and requires no quencher for activation (19). Oxidation is accomplished through a short-lived ligand radical; hydroxyl radicals or other reactive oxygen species are not generated with photoactivation. Guanine damage has been generated in HeLa cells and mitochondria treated with [Rh(phi)2bpy]3+ and light (27, 28). In this experiment, with or without Rh, bacterial cultures are grown to mid-exponential phase, and samples from each culture are irradiated with light. Total RNA is extracted from each sample, and the soxS transcript is amplified using reverse transcription PCR, as is the 23S ribosomal RNA as an internal control.
Fig. 3 shows the results of this experiment. First, the gel shows similar amounts of the 23S RNA PCR product for each sample, indicating that the bacteria are expressing comparable amounts of total RNA. The untreated sample, the sample containing [Rh(phi)2bpy]3+ and not irradiated (DC), and the sample lacking the photooxidant but irradiated (LC), do not show significant soxS expression. However, samples containing [Rh(phi)2bpy]3+ show evidence of strong soxS expression upon irradiation. Under these irradiation conditions, and estimating an intracellular [Rh(phi)2bpy]3+concentration of 5 μM, 1 oxidation event to generate guanine radicals is expected to occur every 300 bp; note that only some of the guanine radicals will lead to irreversible guanine lesions, such as 8-oxo-guanine. Nonetheless, since the Rh photooxidant preferentially causes DNA damage (19), these results indicate that SoxR can be activated in cells by DNA oxidation. Rather than reactive oxygen intermediates inducing SoxR directly, it appears that guanine radicals in DNA can trigger its activation.
Fig. 3.
SoxS expression in E.coli induced by treatment with [Rh(phi)2bpy]3+, a DNA photooxidant, or with methyl viologen. (Upper) An agarose gel image of RT-PCR products stained with ethidium is shown. The top band is a 23S internal control. The bottom band is the soxS transcript. The lowest molecular weight band is free primer. Cells were grown in the presence of 50 μM [Rh(phi)2bpy]Cl3or 50 μM methyl viologen. The asterisk (*) indicates cells that were grown without [Rh(phi)2bpy]Cl3 and not irradiated. LC and DC indicate, respectively, samples irradiated in the absence of metal or treated with metal but not irradiated. The lane labeled 30 represents a culture grown in the presence of [Rh(phi)2bpy]3+ and irradiated for 30 min in 24-well plates without shaking. MV indicates a sample treated with 50 μM methyl viologen and shaken continuously for 30 min at 250 rpm. All irradiations were carried out using a solar simulator with a UV filter and a 1-kW Hg/Xe lamp. LC samples were irradiated for 30 min. (Lower) Levels of soxS bands quantified as the ratio of soxS transcript to the 23S internal control. The 1-tailed P values for the 30 min and MV samples are less than 0.001 compared to the LC. Error bars represent the standard error of the average of 4 separate trials.
Also shown is a comparison of the levels of SoxR activation by Rh photoactivation with that by an equal concentration of methyl viologen, a commonly used activator of SoxR (Fig. 3) (2). It should be noted that the methyl viologen-treated samples were tested aerobically with active shaking of the cultures, while the Rh-treated samples were irradiated in 24-well plates without aeration. Despite these different conditions, the expression levels of soxS in the samples treated with Rh are quite close to those found upon incubation of this E. coli strain with methyl viologen. Note that SoxR is not overexpressed in this wild-type strain. Thus both methyl viologen and the Rh photooxidant, which can activate SoxR only through intermediate DNA oxidation, promote soxS expression to a similar extent.
Transcriptional Activation at a Distance Through DNA-Mediated Oxidation of SoxR.
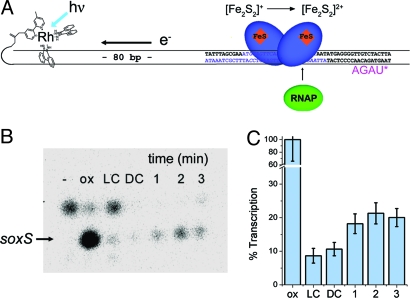
A transcription assay (5) in which SoxR is oxidized from a distance through DNA-mediated charge transport provides a direct test of whether SoxR can be activated in a DNA-mediated reaction. Oxidized SoxR is a powerful transcriptional activator, while reduced SoxR is known to be transcriptionally silent (1, 2). Here, guanine residues on a 180-mer of DNA were oxidized from a distance using [Rh(phi)2bpy′]3+, the intercalating photooxidant, tethered to the end of the DNA fragment (Fig. 4). The 180-bp transcription template used here contains the SoxR binding site and the −10 and −35 promoter regions of soxS, the target gene of SoxR. [Rh(phi)2bpy′]3+ is covalently attached to the DNA at the 5′ end of the duplex (19); at these DNA concentrations, the Rh complex intercalates only intraduplex. Transcription by SoxR was measured in an anaerobic chamber using an abortive transcription assay (29).
Fig. 4.
Transcriptional activation of SoxR from a distance triggered by photoactivation of [Rh(phi)2bpy′]3+ tethered to a 180-mer DNA duplex containing the soxS promoter region. (A) Schematic of the SoxR/DNA complex used to carry out the abortive transcription assay. The SoxR binding site is shown in blue. All experiments were carried out anaerobically. Photooxidation of DNA by the covalently tethered [Rh(phi)2bpy′]3+ oxidizes reduced SoxR bound to DNA. The samples were then incubated with RNA polymerase, and transcription was initiated by addition of a buffer containing a starting dinucleotide, [ApG], ATP, and radiolabeled UTP. The resulting mRNA product is the 4-mer RNA radiolabeled as indicated by the asterisk. Reinitiation of transcription is inhibited by the presence of heparin in the buffer. DNA (30 nM) and 300 nM SoxR are used. Both E. coli and P. aeruginosa SoxR were used (see Methods). P. aeruginosa SoxR was found to show greater stability. (B) Denaturing PAGE of mRNA products of transcriptional activation by SoxR. Shown are the products formed in the absence of SoxR, “-”; in the presence of aerated, oxidized SoxR, “ox”; the DNA template lacking the covalently tethered [Rh(phi)2bpy′]3+ but with reduced SoxR and irradiated, “LC”; the sample with reduced SoxR and covalent Rh/DNA but without irradiation, “DC”; or with reduced SoxR and covalent Rh/DNA and irradiated for increasing time in minutes, 1, 2, 3; samples treated for longer times gave variable precipitation. The top band reflects background transcription and is inhibited by DNA-bound SoxR; the bottom band is the soxS 4-mer transcript. Samples were irradiated in an anaerobic glovebox with a fiber optic cable attached to an LCD lamp (350 nm) (see Methods). (C) Quantitation of soxS expression is given as the percent transcription found versus that with fully oxidized SoxR; data were normalized to the background activity in the dark control (DC). The values for the irradiated samples have 1-tailed P values versus the light control of 0.106, 0.060, and 0.052 for 1, 2, and 3 min, respectively. The error bars represent the standard error of the average of 4 separate trials.
As evident in Fig. 4, in this transcription assay, 2 product bands are seen. The higher molecular weight band is evident in the absence of SoxR and corresponds to background transcription. With SoxR bound to the promoter, this background activity is diminished; we also consistently observe greater basal activity in the light control, which may reflect some SoxR decomposition with irradiation. The band corresponding to the 4-mer transcript of interest is seen most intensely in the control sample containing oxidized SoxR. Samples containing reduced SoxR, which are irradiated in the absence of metal (LC), or samples containing metal and reduced SoxR, which were not irradiated (DC), do not cause oxidative damage to DNA, and in this case, do not cause appreciable transcriptional activation; the amount of transcription is comparable to the background in the gel shown. However, when these anaerobic samples containing reduced SoxR and the tethered metal/DNA complex are irradiated, as shown in the time course averaged over several trials, greater amounts of transcription are observed. Importantly, we cannot detect direct photooxidation of SoxR by the Rh complex without DNA; reduced SoxR incubated with [Rh(phi)2bpy′]3+ at equimolar concentrations but without DNA shows no evidence of SoxR oxidation spectroscopically.
These results indicate that, in an in vitro assay, SoxR oxidized from a distance through DNA-mediated charge transport is transcriptionally active and can express its target gene. The amount of transcriptional activation seen here is small compared to that seen with fully oxidized SoxR, however. This yield is expected given the short-lived excited state of the Rh photooxidant and its low quantum yield of damage; we have seen that the quantum yield for guanine damage is limited by rapid back electron transfer to the long-lived guanine radical (30). Nonetheless, in the abortive transcription assay used, the amount of mRNA transcript produced is a direct measure of the population of oxidized SoxR generated under these conditions of irradiation and corresponds well with the concentration of guanine radicals we may expect to have formed. In a cell, SoxR bound to its promoter region would initiate multiple rounds of transcription, thus amplifying the initial damage signal many times. It is also interesting to note that these results indicate transcriptional activation from a distance 270 Å away. DNA charge transport provides a means to carry out redox chemistry at long range.
Discussion
These experiments provide one answer as to how SoxR is able to sense oxidative stress and activate the response needed to remediate it while bound to a single site on the genome. The first step in this pathway would be the generation of oxidative radicals in DNA by a host of reactive oxygen species that may be present in the cell. Generated radicals would rapidly migrate to areas in the genome of low oxidative potential, such as guanine multiplets, which are found in abundance near the SoxR binding region (5). Moreover, these 1-electron oxidation events in DNA can signal SoxR to become transcriptionally active; DNA-mediated oxidations are exceedingly rapid (ps/ns) if the species to be oxidized is well-coupled into the base stack (16). Thus oxidative guanine radicals that normally react on a slow (ms) timescale to form permanent oxidation products are instead quickly filled by the donation of an electron from reduced SoxR. That these reactions are efficient is evident where the E. coli cultures grown in the presence of a known DNA-binding photooxidant and exposed to light induce the expression of soxS to an extent comparable to aerated samples with methyl viologen. Importantly, we also show here directly that the covalently tethered photooxidant that injects electron holes into the DNA base pair stack can induce the oxidation of SoxR through DNA from a distance to activate transcription. Indeed, these studies present direct evidence of transcriptional activation from a distance.
Oxidative stress, in the form of reactive oxygen species or other oxidative insults, threatens cell survival, and is implicated in DNA damage, aging, and cancer. It is crucial that cells are able to detect and respond to conditions of oxidative stress. These results underscore how the DNA duplex can serve as a conduit to signal information regarding cellular stress across the genome. We have identified other examples of DNA-mediated signaling of oxidative stress: (i) the DNA-mediated oxidation and dissociation of p53 from promoter sites (23) and (ii) the activation of base excision repair proteins by guanine radicals (22). All these examples illustrate how DNA charge transport can effect redox chemistry without the need for co-localization of redox partners. DNA charge transport chemistry offers a remarkably effective and unique means to achieve long range signaling. Other opportunities where this redox chemistry is used within the cell either for transcriptional activation or other regulatory functions should now be considered.
Methods
Attenuation of Oxidative Guanine Products by SoxR.
DNA was synthesized using standard phosphoramidite chemistry on an Applied Biosystems 394 DNA synthesizer (ABI; Glen Research) and HPLC-purified. The sequences prepared were 5′-TTTTCCAATTTTATAAATCGCTTTACCTCAAGTTAACTTGAGGAATTA-3′ and its complement. The complement strand was 5′[32−P]-labeled using polynucleotide kinase and PAGE-purified. [Ru(phen)(dppz)(bpy′)]Cl2 was synthesized using general procedures (31, 32). E. coli SoxR was expressed and purified as outlined (33). DNA (0.5 μM DNA) in 15 mM NaPi, 50 mM NaCl, pH 7.4 was incubated with 5 μM [Ru(phen)(dppz)(bpy′)]2+ and 500 μM Co(NH3)5Cl2+. Experiments were carried out in an anaerobic chamber (Coy). Oxidized or reduced SoxR in 75 mM KCl, 10 mM Tris, 2 mM MgCl2, 10% glycerol, pH 8.0, was added in the indicated concentrations. SoxR was reduced by the addition of a 10-fold excess of sodium dithionite. Complete reduction was checked via UV-Vis spectroscopy (5). Samples were irradiated in anaerobic NMR tubes (J. Young) for 20 min at 442 nM on a He/Cd laser (LiCONiX). After irradiation, DNA was treated with 10% piperidine for 30 min at 90 °C, dried under rotary evaporation, and electrophoresed through a 20% denaturing polyacrylamide gel at 90 W. Gels were visualized via autoradiography using a Storm 820 PhosphorImager (Molecular Dynamics/GE Healthcare) and quantified using ImageQuant (Molecular Dynamics).
Gene Activation in E. coli.
E. coli strain K-12 TB1 (New England Biolabs) was streaked to a Luria-Bertani (LB) agarose plate, and a starter culture was grown from a single colony. A 50-μL aliquot of this starter culture was used to inoculate 5 mL LB or LB containing 50 μM [Rh(phi)2bpy]3+. [Rh(phi)2bpy]Cl3 was prepared as described (34). Cells were grown for 4–5 h until mid-exponential phase, then were pelleted and resuspended in NCE. Aliquots (1 mL) were irradiated in 24-well plates. Irradiations were carried out using a solar simulator (Oriel Instruments) using a 340-nm internal low pass filter and a 320-nm external low pass filter. Samples containing methyl viologen were shaken for 30 min in 1 mL NCE media containing 50 μM methyl viologen. Immediately after irradiation, cells were frozen in liquid N2. Samples were thawed at 60 °C, and RNA was extracted using a RNeasy kit (Qiagen), during which an on column DNase digestion was performed (Qiagen). Reverse transcription was carried out using the OmniScript system (Qiagen), and the resulting cDNA was PCR-amplified using standard procedures. The primers used for RT-PCR were 5′-TTACAGGCGGTGGCGATAAT-3′ and 5′-atgtcccatcagaaaattat-3′ (IDT). Samples were run on a 1.3% agarose gel and stained with ethidium bromide. The gel was visualized under a UV transilluminator and photographed.
Abortive Transcription Assay.
[Rh(phi)2bpy′]Cl3 was synthesized as described (35). Primers A (5′-CTGAATAATTTTCTGATGGG-3′) and B (5′-GCCACACCGCTGCGTTTCGC-3′) were synthesized as described above. Primer B was covalently modified with the photooxidant [Rh(phi)2bpy′]3+ as described previously (36). Tethering was confirmed by MALDI-mass spectrometry. Primers A and B were used to PCR amplify a 180-bp fragment of DNA containing the SoxR binding site and the start of the soxS gene from E. coli genomic DNA. The abortive transcription assay was modified from one published previously (5). DNA (30 fmol in 15 mM NaPi, 50 mM NaCl, pH 7.4) was incubated in 30 μL 300 nM SoxR (75 mM KCl, 10 mM Tris, 2 mM MgCl2, pH 8.0), from E. coli or Pseudomonas aeruginosa (32, 12). Reduced samples were prepared as described previously using dithionite in an anaerobic chamber. The samples were then irradiated in the glovebox using a Schott KL2500 LCD lamp outfitted with the orange filter and at the highest power and iris settings. RNA polymerase (1/25 A.U.; Epicentre) was added in a 2-μL volume, and allowed to incubate for 15 min. A 10-μL aliquot of this mixture was removed from each sample and replaced with 10 μL transcription activation buffer [6 mM ApG (Dharmacon), 0.1 mM ATP, 0.25 μM [α-32P]UTP (>600 Ci/mmol; MP Biomedical), 0.3 M K-glutamate, 30 mM Tris-HCl, pH 8.0, 3 mM MgCl2, 0.3 mg/mL BSA, 3 mM CaCl2], and samples were allowed to incubate in the glovebox for 1 h at 37 °C. Reactions were quenched using 3 μL 50 mM EDTA, 30% glycerol, and 8 μL formamide loading buffer. Aliquots (8 μL) were loaded onto a 20% denaturing polyacrylamide gel and were electrophoresed at 90 W for 90 min. The gels were visualized using standard autoradiography as described above.
Acknowledgments.
We thank the National Institutes of Health (NIH) (GM49216 to J.K.B. and CA37831 to B.D.) for their financial support of this research. We thank also Dr. Eunsuk Kim and Dr. Lars Dietrich for their preparations of E. coli and P. Aeruginosa SoxR, respectively.
Footnotes
The authors declare no conflict of interest.
References
- 1.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amábile-Cuevas CF, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: Two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaudu P, Moon N, Weiss B. Regulation of the soxRS oxidative stress regulon. Reversible oxidation of the Fe-S centers of SoxR in vivo. J Biol Chem. 1997;272:5082–5086. doi: 10.1074/jbc.272.8.5082. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, Dunham WR, Weiss B. Overproduction and physical characterization of SoxR, a [2Fe-2S] protein that governs an oxidative stress response regulon in Escherichia coli. J Biol Chem. 1995;270:10323–10327. doi: 10.1074/jbc.270.17.10323. [DOI] [PubMed] [Google Scholar]
- 5.Hidalgo E, Demple B. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 1994;13:138–146. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hidalgo E, Demple B. Activation of SoxR-dependent transcription in vitro by non-catalytic or NifS-mediated assembly of [2Fe2S] clusters into apo-SoxR. J Biol Chem. 1996;271:7269–7272. doi: 10.1074/jbc.271.13.7269. [DOI] [PubMed] [Google Scholar]
- 7.Ding H, Hidalgo E, Demple B. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J Biol Chem. 1996;271:33173–33175. doi: 10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- 8.Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci USA. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hidalgo E, Demple B. Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. EMBO J. 1997;16:1056–1065. doi: 10.1093/emboj/16.5.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe S, Kita A, Kobayashi K, Miki K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc Natl Acad Sci USA. 2008;105:4121–4126. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobman J. MerR family transcription activators: Similar designs, different specificities. Mol Microbiol. 2007;63:1275–1278. doi: 10.1111/j.1365-2958.2007.05608.x. [DOI] [PubMed] [Google Scholar]
- 12.Gorodetsky A, et al. DNA binding shifts the redox potential of the transcription factor SoxR. Proc Natl Acad Sci USA. 2008;105:3684–3689. doi: 10.1073/pnas.0800093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutton V, et al. Superoxide destroys the [2Fe-2S] Cluster of FNR from Escherichia coli. Biochemistry. 2009;43:791–798. doi: 10.1021/bi0357053. [DOI] [PubMed] [Google Scholar]
- 14.Machado M, Michán C, Pueyo C. Hydrogen peroxide activates the SoxRS regulon in vivo. J Bacteriol. 2000;182:6842–6844. doi: 10.1128/jb.182.23.6842-6844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beckman KB, Ames BN. Oxidative decay of DNA. J Biol Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 16.Delaney S, Barton JK. Long-range DNA charge transport. J Org Chem. 2003;68:6475–6483. doi: 10.1021/jo030095y. [DOI] [PubMed] [Google Scholar]
- 17.Burrows CJ, Muller JG. Oxidative nucleobase modifications leading to strand scission. Chem Rev. 1998;98:1109–1151. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 18.Merino EJ, Boal AK, Barton JK. Biological contexts for DNA charge transport chemistry. Curr Opin Chem Biol. 2008;12:229–237. doi: 10.1016/j.cbpa.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall DB, Holmlin RE, Barton JK. Oxidative DNA damage through long-range electron transfer. Nature. 1996;382:731–735. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- 20.Boon EM, Salas JE, Barton JK. An electrical probe of protein-DNA interactions on DNA-modified surfaces. Nat Biotechnol. 2002;20:282–286. doi: 10.1038/nbt0302-282. [DOI] [PubMed] [Google Scholar]
- 21.Nunez ME, Hall D, Barton JK. Long-range oxidative damage to DNA: Effects of distance and sequence. Chem Biol. 1999;6:85–97. doi: 10.1016/S1074-5521(99)80005-2. [DOI] [PubMed] [Google Scholar]
- 22.Yavin E, et al. Long-range electron transfer special feature: Protein-DNA charge transport: Redox activation of a DNA repair protein by guanine radical. Proc Natl Acad Sci USA. 2005;102:3546–3551. doi: 10.1073/pnas.0409410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Augustyn KE, Merino EJ, Barton JK. A role for DNA-mediated charge transport in regulating p53: Oxidation of the DNA-bound protein from a distance. Proc Natl Acad Sci USA. 2007;104:18907–18912. doi: 10.1073/pnas.0709326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stemp EDA, Arkin MR, Barton JK. Oxidation of guanine in DNA by Ru(phen)2(dppz)3+ using the flash-quench technique. J Am Chem Soc. 1997;119:2921–2925. [Google Scholar]
- 25.Arkin MR, Stemp EDA, Barton JK. Long-range oxidation of guanine by Ru(III) in duplex DNA. Chem Biol. 1997;4:389–400. doi: 10.1016/s1074-5521(97)90129-0. [DOI] [PubMed] [Google Scholar]
- 26.Saito I, et al. Photoinduced DNA cleavage via electron transfer: Demonstration that guanine residues located 5′ to guanine are the most electron-donating sites. J Am Chem Soc. 1995;117:6406–6407. [Google Scholar]
- 27.Nunez ME, Holmquist GP, Barton JK. Evidence for DNA charge transport in the nucleus. Biochemistry. 2001;40:12465–12471. doi: 10.1021/bi011560t. [DOI] [PubMed] [Google Scholar]
- 28.Merino EJ, Barton JK. DNA oxidation by charge transport in mitochondria. Biochemistry. 2008;47:1511–1517. doi: 10.1021/bi701775s. [DOI] [PubMed] [Google Scholar]
- 29.Rhodius V, et al. Methods in Molecular Biology. 2nd Ed. vol 148. Totawa, NJ: Humana Press; 2001. pp. 451–464. [Google Scholar]
- 30.Williams T, Dohno C, Stemp ED, Barton JK. Effects of the photooxidant on DNA-mediated charge transport. J Am Chem Soc. 2004;126:8148–8158. doi: 10.1021/ja049869y. [DOI] [PubMed] [Google Scholar]
- 31.Strouse GF, Anderson PA, Schoonover JR, Meyer TJ, Keene FR. Synthesis of polypyridyl complexes of ruthenium(II) containing three different bidentate ligands. Inorg Chem. 1992;31:3004–3006. [Google Scholar]
- 32.Anderson PA, et al. Designed synthesis of mononuclear Tris(heteroleptic) ruthenium complexes containing bidentate polypyridyl ligands. Inorg Chem. 1995;34:6145–6157. [Google Scholar]
- 33.Chandler M, Demple B. Functional analysis of SoxR residues affecting transduction of oxidative stress signals into gene expression. J Biol Chem. 2004;279:41603–41610. doi: 10.1074/jbc.M405512200. [DOI] [PubMed] [Google Scholar]
- 34.Sitlani A, Barton JK. Sequence-specific recognition of DNA by phenanthrenequinone diimine complexes of rhodium(III): Importance of steric and van der Waals interactions. Biochemistry. 1994;33:12100–12108. doi: 10.1021/bi00206a013. [DOI] [PubMed] [Google Scholar]
- 35.Pyle AM, Chiang MY, Barton JK. Synthesis and characterization of physical, electronic, and photochemical aspects of 9,10-phenanthrenequinonediimine complexes of ruthenium(II) and rhodium(III) Inorg Chem. 1990;29:4487–4495. [Google Scholar]
- 36.Holmlin RE, Dandliker PJ, Barton JK. Synthesis of metallointercalator-DNA conjugates on a solid support. Bioconjugate Chem. 1999;10:1122–1130. doi: 10.1021/bc9900791. [DOI] [PubMed] [Google Scholar]