Figure 1.
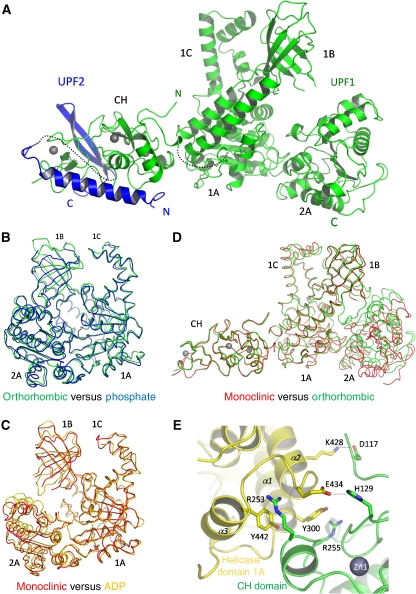
Structure of the UPF1(115–914)–UPF2(1105–1198) complex. (A) Ribbon diagram of the complete structure, with UPF1 in green and UPF2 in blue. The missing links between the UPF1 CH- and helicase domains and between the N and C-terminal parts of UPF2 are represented as dotted lines. (B) Superposition of the closed form of the helicase domain (orthorhombic crystal, green) with the previously described helicase domain in the phosphate-bound form (PDB ID 2gk7, blue). The RMSD between the two structures is 1.03 Å for 591 aligned Cα atoms. The RMSD values between the orthorhombic form and the AMPPNP (PDB ID 2gjk) and ADP (PDB ID 2gk6) forms are, respectively, 1.81 and 2.00 Å. (C) Superposition of the open form of the helicase domain (monoclinic crystal, red) with the previously described helicase domain in the ADP-bound form (PDB ID 2gk6, gold). The RMSD between the two structures is 1.35 Å for 584 aligned Cα atoms. The RMSD values between the monoclinic form and the phosphate and AMPPNP forms are, respectively, 2.48 and 3.07 Å. (D) Superposition of UPF1 from the monoclinic (red) and orthorhombic (green) crystal forms showing that the relative orientations of the CH and helicase domains are the same in each case, although the helicase conformation is different. (E) The principal interacting residues from the CH- (green) and helicase (yellow) domains of UPF1 are represented as sticks. The same interactions are found in both monoclinic and orthorhombic crystal forms. These residues are well conserved (Supplementary Figure S2).