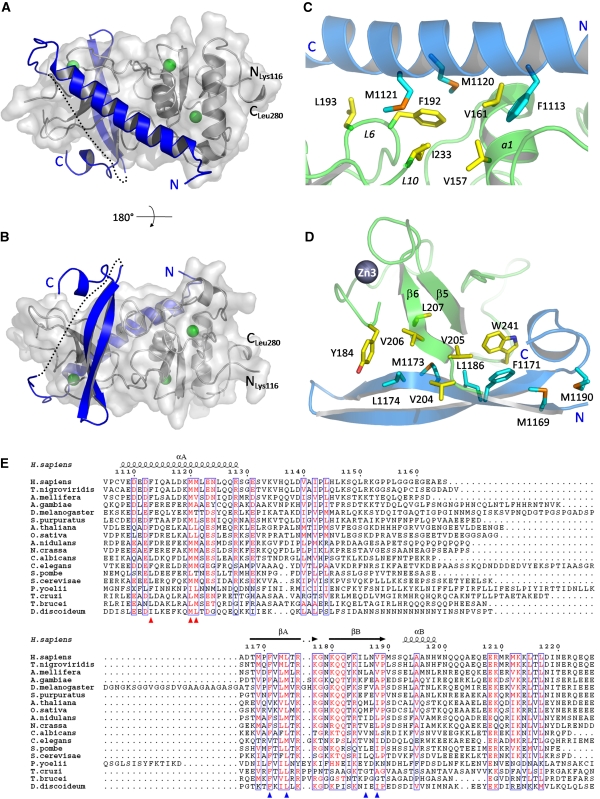
Figure 2.
UPF2 binds on two opposite surfaces of the UPF1 CH domain. (A, B) UPF2 (blue) is represented as ribbons and UPF1 (grey) as ribbons and a transparent surface. UPF1 zinc atoms are shown in green. The UPF2 missing linker is represented as a dotted line. The two views differ by a rotation of 180 degrees around the horizontal axis. (C) The principal residues of the UPF2 N-terminal helix (cyan) and the UPF1 CH-domain (yellow), which form the hydrophobic interface between the two molecules, are represented as sticks. (D) The main interacting residues of UPF2 C-terminal β-hairpin (cyan) and UPF1 CH domain (yellow) are represented as sticks. Met 1169 and Met 1190 do not interact directly with UPF1 binding but form part of a small hydrophobic core important for the stability of the bound form of UPF2. (E) Sequence alignment of the UPF1-binding domain of representative UPF2 proteins from yeast to human. Residues with similarity >70% are displayed in red. The secondary structure of the UPF1-bound human UPF2 is indicated as α (alpha-helix) and β (beta-strand). Red and blue triangles indicate the main UPF1-interacting residues belonging to the N-terminal helical region and the C-terminal β-hairpin, respectively. The alignment was generated with ClustalX (Thompson et al, 2002) and showed using ESPript (http://espript.ibcp.fr/ESPript/ESPript/).