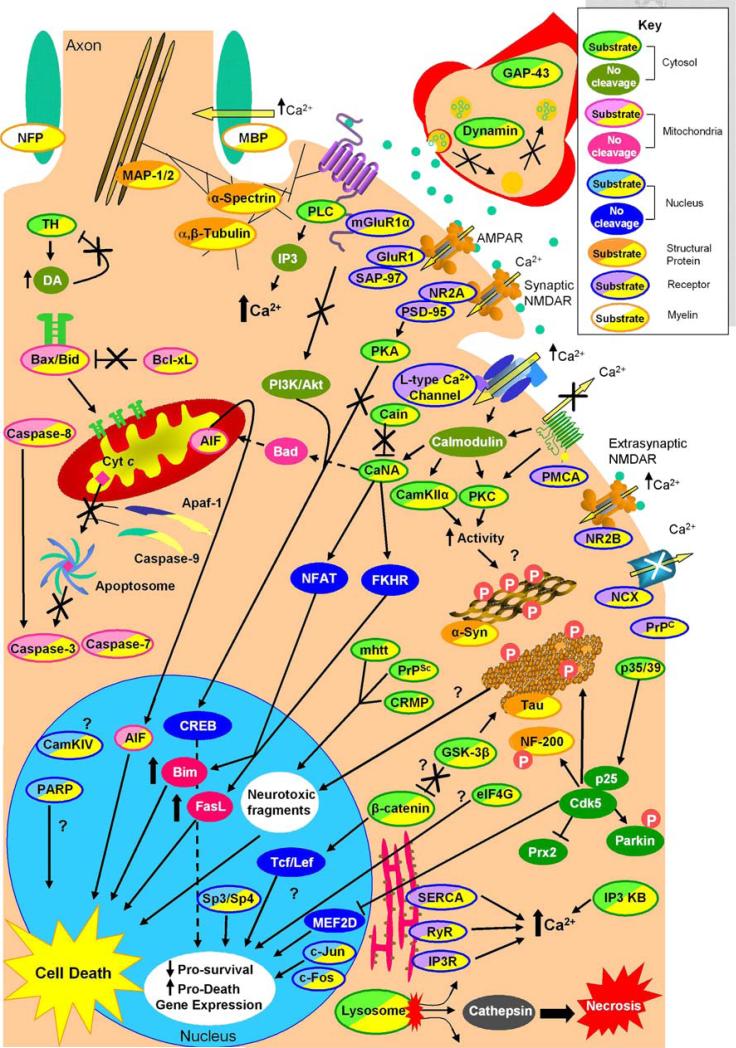
Fig. 2.
Intracellular signaling consequences of calpain-mediated substrate cleavage. Examination of the effects of calpain cleavage on its numerous substrates can be categorized by subcellular localization or protein class. Here, calpain substrates are divided into receptor, cytosolic, mitochondrial, nuclear, structural, and myelin proteins (see key). Intracellular calcium concentration is tightly regulated by membrane and endoplasmic reticulum associated receptors. Calpain propagates its own activation by blocking the calcium purging actions of PMCA and NCX, increasing calcium influx via l-type calcium channel cleavage and causing ER release of calcium via cleavage of SERCA, RyR, and IP3R. Cleavage of mGluR1α and the NMDAR NR2A subunit the C-termini uncouples both receptors from prosurvival CREB activation and potentiates increased calcium levels via increased PLC activity. Cleavage of calcium-regulated proteins such as PKC and CamKIIα also increase their activity to potentially hyperphosphorylate and cause aggregation of α-synuclein and tau. CaN A cleavage increases its activity causing dephosphorylation and translocation of transcription factors NFAT and FKHR leading to increased Bim and FasL levels. Cleavage of p35 to p25 alters Cdk5 activity, which is also involved with hyperphosphorylation of α-synuclein and tau, and also translocates to the nucleus and turns off prosurvival gene expression via MEF2D and increases ROS by inhibition of Prx2. The substrate β-catenin may also be involved with calpain induced cell death as cleavage mediates gene transcription. Finally, in the cytosol, calpain cleavage products from proteins such as mhtt, CRMP, PrPSc, and potentially tau translocate to the nucleus and are neurotoxic. Calpain also has substrates involved with mitochondria-associated programmed cell death. Calpain directly cleaves most of the caspases, Bax and Bcl-xL, and AIF, and it indirectly activates Bad via increased CaN A activation to initiate programmed neuronal death. However, calpain can also inactivate this pathway by cleaving Apaf-1 and caspase-9 in a model-dependent manner. Calpain is also active in the nucleus where it cleaves CamKIV and PARP, the transcription factors Sp3, Sp4, and the immediate early genes c-Jun and c-Fos. The effect of CamKIV and PARP cleavage has not been delineated. Sp3, Sp4, c-Fos, and c-Jun cleavage is thought to decrease housekeeping gene transcription leading to cell death; however, c-Jun-mediated transcription has also bee implicated in cell death. Lastly, structural proteins such as α-spectrin, MAP-1/2, and α- and β-tubulin are calpain substrates. Cleavage of these proteins is thought to impair microtubule transport and cell body and synaptic neuronal structures, potentially leading to neuronal death. α-Syn α-Synuclein, AIF apoptosis-inducing factor, AMPAR alpha-amino-3hydroxy-5-methyl-4-isoxazolepropionic acid receptor, Apaf-1 apoptosis-activating factor 1, CAMKIIα Ca2+/calmodulin-dependent protein kinase IIα, CAMKIV Ca2+/calmodulin-dependent protein kinase IV, CaN A calcineurin A, Cdk5 cyclin-dependent kinase 5, CREB cyclic AMP response element binding protein, CRMP collapsing response mediator protein, Cyt c cytochrome c, DA dopamine, eIF4G eukaryotic initiation factor 4G, ER endoplasmic reticulum, FasL Fas ligand, FKHR forkhead in rhabdomyosarcoma, GAP-43 growth-associated protein-43, GluR1 AMPA receptor 1, GSK-3β glycogen synthase kinase 3β, IP3 inositol triphosphate, IP3 KB inositol 1,4,5 triphosphate kinase B, IP3R inositol 1,4,5 triphosphate receptor, MAP1,2 microtubule-associated protein 1, 2, MBP myelin basic protein, mhtt mutant huntingtin, mGluR1α metabotropic glutamate receptor, NFP axonal neurofilament protein, NCX sodium–calcium exchanger, NF 200 neurofilament protein 200, NFAT nuclear factor activating T-cells, NMDA NR2A N-methyl-d-aspartate receptor NR2A subunit, NMDA NR2B N-methyl-d-aspartate receptor NR2B subunit, PI3K/Akt phosphatidyl inositol 3 kinase/protein kinase B, PLC phospholipase C, PMCA plasma membrane Ca2+ ATPase, PARP poly (ADP-ribose) polymerase, PKA protein kinase A, PKC protein kinase C, PrPC prion-related peptide, PrPSc scrapie form of prion-related protein, PSD-95 postsynaptic density-95 protein, RyR ryanodine receptor, SAP-97 synapse-associated protein-97, SERCA sarcoplasmic/endoplasmic reticulum calcium ATPase, TH tyrosine hydroxylase