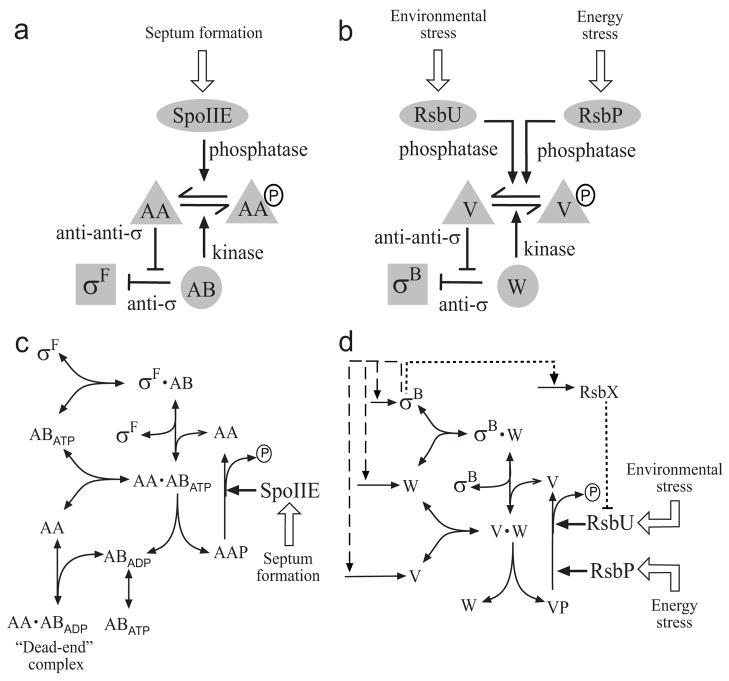
Fig. 1.
Conceptual (a, b) and kinetic (c, d) models of the partner-switching networks regulating σF and σB. For simplicity, only one subunit of the anti-sigma factor dimer is shown, and binding of sigma to RNA polymerase core is omitted as it does not affect the partner-switching equilibrium (see Modeling Procedures). (a) Level of free σF is determined by the level of unphosphorylated SpoIIAA anti-anti-sigma (AA), where P indicates serine phosphate (see text). Activity of the SpoIIE phosphatase in the forespore increases upon formation of the asymmetric sporulation septum, turning the system ON. (b) Level of free σB is determined by the level of unphosphorylated RsbV anti-anti-sigma (V). Activity of either the RsbU environmental phosphatase or the RsbP energy phosphatase increases when a stress is perceived, turning the system ON. (c) Formation of a long-lived, dead-end complex in the σF network. The SpoIIAB anti-sigma factor (AB) with ATP in its catalytic site can phosphorylate and release the AA anti-anti-sigma factor, leaving AB associated with ADP in a non-catalytic complex. The binding of another AA molecule locks AB in this state. (d) Feedback loops in the σB network. Genes encoding σB, its anti-sigma RsbW (W) and anti-anti-sigma V lie in the σB-controlled sigB operon. The resulting feedback loops are shown by dashed lines. An additional feedback loop through the RsbX phosphatase, which acts indirectly via the environmental-stress branch of the network, is shown by the dotted line. Each of the nodes containing W and its complexes are assumed to be in equilibrium, with a mixed population of ATP, ADP and nucleotide-free forms, so no explicit nucleotide exchange is shown.