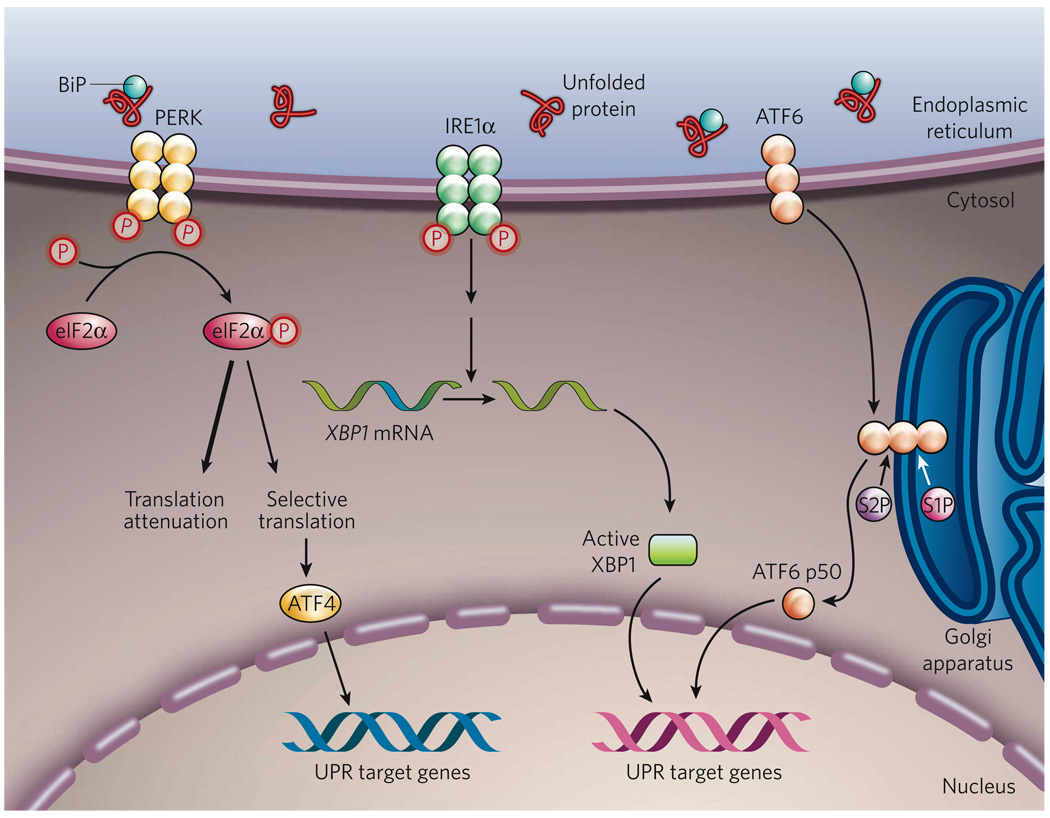
Figure 1. The mammalian UPR pathways.
In non-stressed cells (not shown), the ER chaperone BiP binds to the luminal domains of the ER-stress sensors IRE1α, PERK and ATF6, maintaining these proteins in an inactive state. During ER stress (shown), BiP preferentially binds to unfolded or misfolded proteins, thus driving the equilibrium of BiP binding away from IRE1α, PERK and ATF6. These three proteins are the initiators of the three main signalling cascades of the UPR. The release of BiP results in the activation of PERK, through PERK homodimerization and trans-autophosphorylation. Activated PERK then phosphorylates the translation-initiation factor eIF2α, reducing the overall frequency of messenger RNA translation initiation. However, selected mRNAs, such as ATF4 mRNA, are preferentially translated in the presence of phosphorylated eIF2α. ATF4 activates the transcription of UPR target genes encoding factors involved in amino-acid biosynthesis, the antioxidative-stress response and apoptosis. The release of BiP also allows IRE1α to dimerize, activating its protein-kinase activity (through autophosphorylation) and its endoribonuclease activity. IRE1α then removes a 26-base intron from XBP1 mRNA. The spliced XBP1 mRNA encodes a potent transcription factor that translocates to the nucleus, activating the expression of UPR target genes. The release of BiP from ATF6 allows ATF6 to translocate to the Golgi apparatus, where it is cleaved by the proteases S1P and S2P, yielding an active cytosolic ATF6 fragment (ATF6 p50). This fragment migrates to the nucleus, activating the transcription of UPR target genes. S1P, site-1 protease; S2P, site-2 protease; XBP1, X-box-binding protein 1.