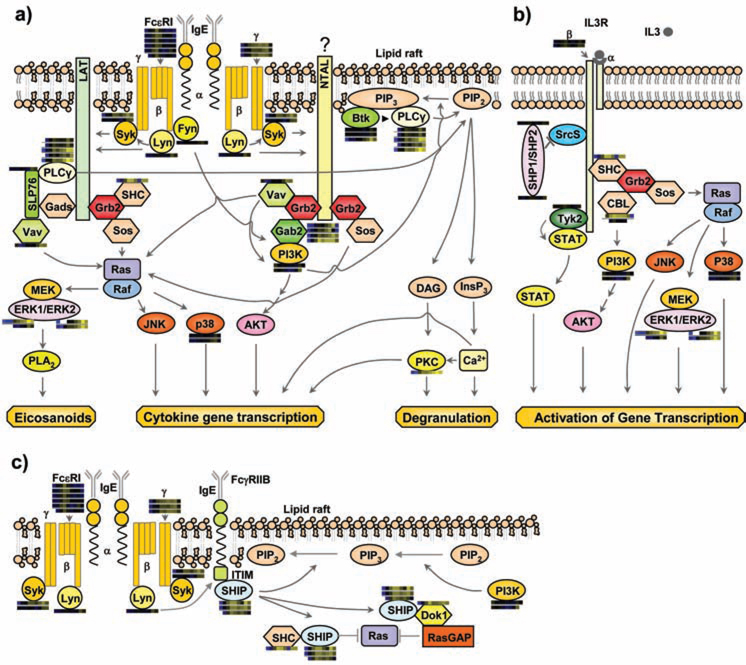
Figure 1.
Multiple previously established signaling cascades in activated mast cells with quantitative proteomic data from MCP5 cells represented as heatmap bars beside individual proteins. These heatmap bars represent the change of abundance of phosphorylation on known mast cell signaling proteins through a timecourse of Fc receptor stimulation (as additionally portrayed in Figure 4a). a) FcεRI-mediated activation pathways. b) IL-3 receptor mediated pathway. c) A representative inhibitory pathway. Syk: spleen tyrosine kinase; LAT: linker for activation of T cells; Grb2: growth-factor-receptor-bound protein 2; Sos: son of sevenless homologue; SHC: SH2-domain-containing transforming protein C; Gads: Grb2-related adaptor protein 2; SLP76: SH2-domain-containing leukocyte protein of 65 kDa; PLCγ: phospholipase C γ; NTAL: non T cell activation linker; Gab2: Grb2-associated binding protein 2; ERK: extracellular-signal-regulated kinase; PLA2: phospholipase A2; p38: Mitogen activated protein kinase 14; PIP2: phosphatidylinositol-4,5-bisphosphate; PIP3: phosphatidylinositol-3,4,5-triphosphate; BTK: bruton's tyrosine kinase; DAG: diacylglycerol; InsP3: inositol-1,4,5-triphosphate; PKC: protein kinase C; SHP1/SHP2: SH2 containing protein tyrosine phosphatase 1/2; Srcs: Src family kinases ; Dok1: docking protein, 62 KD; RasGAP: GTPase activating protein .