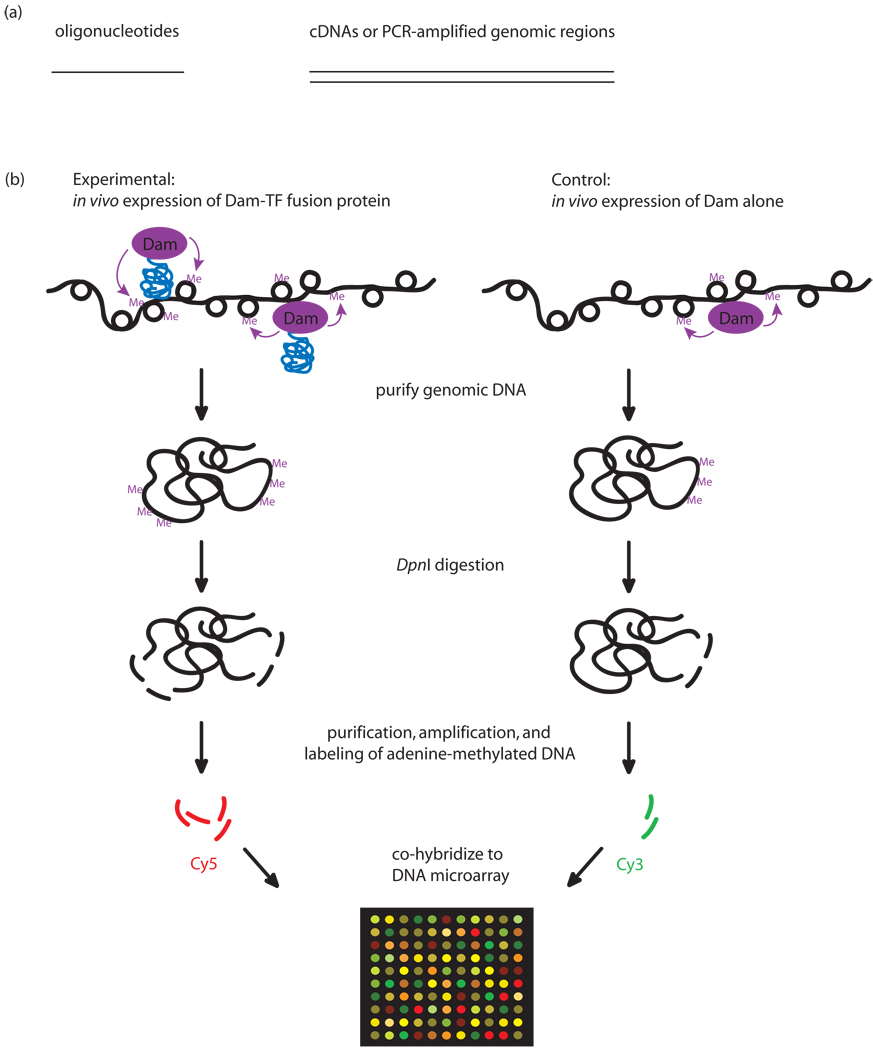
Figure 2. DamID.
(a) Types of DNAs used in DamID microarray hybridizations. Oligonucleotide tiling genomic regions (left); cDNAs or PCR amplification of genomic regions (right). (b) DamID experimental design. The protein of interest (blue) is overexpressed in vivo from a plasmid as a fusion to Dam (purple). Wherever the protein binds DNA, Dam will methylate adenines within GATC sites in the vicinity of the binding sites. The methylated sites are digested with the methyl-specific restriction enzyme DpnI, which cuts only at methylated GATC sites. The smaller DpnI digestion fragments, corresponding to the methylated regions, are either purified by sucrose gradient centrifugation or specifically amplified using a methylation-specific PCR protocol. (Panel (b) was adapted from [7] and [51] with permission from Nature Publishing Group.)