Figure 5.
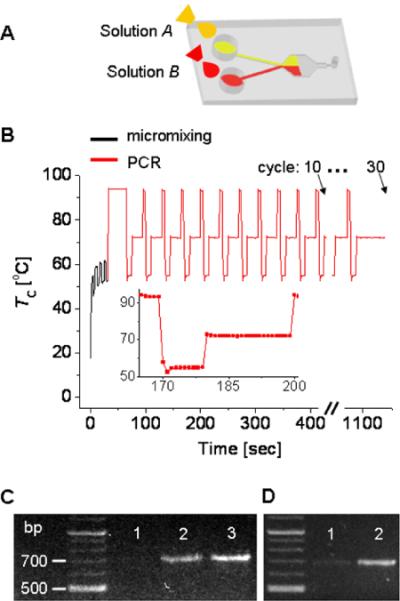
Natural convection-driven micromixing and PCR in a single microfluidic chamber. (A) Pumpless sample transport by capillarity. Solution A is a DNA template and solution B is a reaction mixture of primer, enzyme, and dNTPs. (B) Time sequence of heating used for the AH mode micromixing and the PCR process. The inset shows a single heat cycle of the PCR process with a time interval of 1 sec between the adjacent data points. PCR-based amplification of a DNA fragment from the influenza viral strain A/LA/1/87 is performed for 10, 20, and 30 cycles. (C) Influence of PCR cycles. Lanes 1, 2, and 3 correspond to the amplified PCR products after the microfluidic mixing and the subsequent 10, 20, and 30 PCR cycles, respectively. (D) Improvement of PCR by microfluidic mixing. Both lane 1 and lane 2 are the amplified PCR products after the 20 cycle. However, lane 1 is a control without a microfluidic mixing process at AH mode.
