Abstract
Heat shock protein 90 (HSP90) works as a multi-functional chaperone and is involved in the regulation of many essential cellular pathways. In this study, we have identified a full-length complementary DNA (cDNA) of HSP90 (FcHSP90) from Chinese shrimp Fenneropenaeus chinensis. FcHSP90 full-length cDNA comprised 2,552 bp, including a 2,181-bp open reading frame encoding 726 amino acids. Both homology analyses using alignment with previously identified HSP90 and a phylogeny tree indicated that FcHSP90 was a cytoplasmic HSP90. Real-time reverse transcription polymerase chain reaction analysis revealed that FcHSP90 was ubiquitously expressed in all the examined tissues but with highest levels in ovary of F. chinensis. FcHSP90 mRNA levels were sensitively induced by heat shock (from 25°C to 35°C) and reached the maximum at 6 h during heat shock treatment. Under hypoxia conditions, FcHSP90 mRNA levels, in both hemocytes and gill, were induced at 2 h and depressed at 8 h during hypoxia stress. The assessment of FcHSP90 mRNA levels under heat shock and hypoxia stresses indicated that the transcription of FcHSP90 was very sensitive to heat shock and hypoxia, so we deduced that FcHSP90 might play very important roles for shrimp to cope with environmental stress.
Keywords: Heat shock protein 90, Fenneropenaeus chinensis, Heat shock, Hypoxia
Introduction
Heat-shock proteins (HSPs) are ubiquitous proteins that cope with stress-induced denaturation of client proteins in diverse organisms. Recent studies demonstrated that HSPs display other essential roles including folding, assembly, intracellular localization and degradation of other proteins, and regulation of gene expression (Terasawa et al. 2005). Generally, HSPs are grouped into five major families on the basis of their molecular size in kilodaltons (kDa): HSP110, HSP90, HSP70, HSP60, and the low-molecular weight HSP families. To date, most studies have focused on the HSP70 and HSP90 families. The HSP70 family consists of several members with similar molecular sizes, including constitutively expressed heat shock cognate protein 70 (HSC70), heat-inducible heat shock protein70 (HSP70), glucose-regulated protein GRP78, etc. HSP70 and HSC70 are cytosolic members of the HSP70 family, while GRP78 is located in the endoplasmic reticulum. According to the subcellular location of HSP90, the HSP90 family is classified into four types including cytoplasmic, endoplasmic reticulum, mitochondria, and chloroplast in plants (Picard 2002).
It was already reported that heat shock proteins are rapidly synthesized within stressed cells after exposure to environmental stressors (Bendena et al. 1991; Lang et al. 2000). HSPs can be induced by a variety of environmental factors including heat (Schlesinger 1990; Currie and Tufts 1997, Piano et al. 2005), trace-metal exposure (Sanders et al. 1991; Williams et al. 1996, Schill et al. 2003), organic pollutants (Sanders et al. 1991), anoxia (Myrmel et al. 1994), osmolarity (Kültz, 1996), and Vibrio infection (Cellura et al. 2006).
The aquatic environment is a very complex system in which temperature, salinity, pollutant content, and oxygen will vary greatly depending on the season, weather condition, or human activity. Variations in the aquatic environment will have a great effect on many biological processes of the organism such as development, growth, and reproduction. In this case, HSPs seem to be very important in enabling the animals to cope with the variations of a complex environment. In recent years, increasing interest has been paid to the study of the expression of HSPs in aquatic animals, especially in some aquaculture species. HSP70 genes and their expression have been reported in mollusks such as the European flat oyster, Ostrea edulis (Piano et al. 2005), Mytilus galloprovincialis (Cellura et al. 2006), the bay scallop, Argopecten irradians (Song et al. 2006), the abalone, Haliotis tuberculata (Farcy et al. 2007); in fish such as tilapia Oreochromis mossambicus (Molina et al. 2000); in crustaceans such as Chinese shrimp, Fenneropenaeus chinensis (Jiao et al. 2004), Macrobrachium rosenbergii (Liu et al. 2004), and tiger shrimp Penaeus monodon (Lo et al. 2004).
Compared with the progress made in the study of HSP70, fewer reports about HSP90 have been found in aquatic animals (Palmisano et al. 2000; Deane et al. 2002; Farcy et al. 2007; Gao et al. 2007). Penaeid shrimp are very important in world aquaculture. Studies on HSPs of shrimp can help us to understand the biological process by which shrimp cope with various stresses.
Cytoplasmic HSP90, associated with its co-chaperones containing a conserved tetratricopeptide repeat motif, is involved in cell regulatory pathways by activating hundreds of client proteins including kinases, transcription factors, and steroid receptors (Terasawa et al. 2005; Pearl and Prodromou 2006; Travers and Fares 2007). HSP90 protein structure comprises three conserved domains: the N-terminal domain with adenosine triphosphate (ATP)-binding site (Prodromou et al. 1997), the core domain interacting with some client proteins such as p53 (Muller et al. 2004), and the C-terminal domain responsible for dimerization (Brown et al. 2007). HSP90 generally responds to thermal stress by decreasing the aggregation of denatured proteins or of aggregated proteins for degradation (Palmisano et al. 2000; Picard 2002; Spees et al. 2002; Terasawa et al. 2005). In the present study, a cytoplasmic HSP90 was cloned from Chinese shrimp F. chinensis, and its expression was analyzed by real-time reverse transcription polymerase chain reaction (RT-PCR) under heat shock or hypoxia stress.
Materials and methods
Shrimp source
Shrimp for cDNA cloning and tissue distribution Healthy Chinese shrimp (F. chinensis) with a body weight of 11.5 ± 1.2 g and a body length of 10.2 ± 0.8 cm were collected from a local shrimp farm near Qingdao. The shrimp were acclimated in the aerated seawater at 25 ± 1°C for 5–7 days in the lab before the experiment. The hemolymph was withdrawn from the ventral sinus located at the first abdominal segment using an equal volume of modified Alsevier solution anticoagulant (Bachère et al. 1988) and subsequently centrifuged for 10 min at 800×g, 4°C. Hemocyte pellets were immediately used for RNA extraction. After the hemolymph was collected, different tissues including hepatopancreas, heart, gill, intestine, lymphoid organ, nerve, epidermis, and ovary were dissected out and preserved in liquid nitrogen for later RNA extraction.
Shrimp for heat shock experiment For field culture of F. chinensis, juvenile shrimp usually encounter high temperatures from late July to early August. In order to learn the response of shrimp to heat shock stress, we used juvenile shrimp in the heat shock experiment. Juvenile shrimp of 4–5 cm length, cultured in our lab for 2 months from fertilized eggs, were transferred in our lab to 0.3 m3 containers with seawater at a temperature of 25 ± 1°C (which is approximately the room temperature) for 5 days of acclimation. Six shrimp were randomly selected as 0 h samples before heat shock treatment. The shrimp were removed to another container with seawater maintained at a temperature of 35 ± 1°C for 6 h, and these shrimp were then put back to containers at 25 ± 1°C for a continuous post-stress recovery of 48 h. Six shrimp were randomly collected at 1, 2, 3, 4, and 6 h separately during the heat shock treatment at 35 ± 1°C. After heat shock treatment, the shrimp were transferred from 35 ± 1°C to 25 ± 1°C to be cultured for recovery. During the recovery time, six shrimp were sampled at 6, 24, and 48 h separately. The cephalothorax of the shrimp at each sampling time were separated, frozen immediately in liquid nitrogen, and stored at −80°C until RNA extraction for later gene expression analysis. No mortality was observed during the experiment.
Shrimp for hypoxia exposures During the field shrimp culture, shrimp of larger size coped better with hypoxia stress than juveniles at the same shrimp density. For above reason, we used mature shrimp to do the hypoxia experiments. The gill is the main organ for shrimp to exchange oxygen with sea water, and the oxygen can be carried to the whole body of the shrimp through hemolymph. In order to study the effect of hypoxia on the shrimp, gill and hemocytes are the best tissues for gene expression analysis under hypoxia stress. Seventy healthy Chinese shrimp with 11.0 ± 1.8 cm length and 11.5 ± 1.2 g weight were collected from a local shrimp farm near Qingdao and transferred to the aquarium of our institute. The shrimp were kept in fibril glass tank (its effective volume is about 4 kl) with sea water at about 20°C (room temperature), 30 ppt salinity, for acclimation for 5 days prior to the experiment. After acclimation, six shrimp were collected as the control group before hypoxia treatment. Their hemocytes were collected as described above for RNA extraction. The gills were dissected out and preserved in liquid nitrogen for further RNA analysis. Two tanks with volume of 400 l were prepared with sea water at temperature of 20°C and salinity of 30 ppt. In one tank, normoxia condition was obtained by equilibrating sea water with air of which the concentration of O2 was at 8.5–9.5 mg/l (100% v/v O2 saturation). Ten shrimp were kept in the normoxia condition until the end of the experiment. In another tank, hypoxia conditions were controlled through oxygen removal by nitrogen flow. Before the shrimp were removed to the tank, the O2 concentration of sea water was decreased to 5.30 mg/l (about 60% v/v O2 saturation), then nitrogen flow was stopped. Then 54 shrimp were transferred into the hypoxia sea water from normoxia condition. Due to respiration of shrimp, the O2 concentration of sea water will drop continuously. The concentration of O2 was detected by a portable meter for dissolved oxygen measurement (DO200 model, Lovibond, Germany) every hour. At 2 h post hypoxia (about 50% O2 saturation), six shrimp were randomly taken out to collect their hemocytes and gills for further analysis. At 8 h post hypoxia (about 28% O2 saturation), six shrimp were taken out, and their hemocytes and gills were sampled. The water was then air bubbled to restore the normoxia condition until the next day. The hypoxia condition was set for about 4 h on the next day, then restored to normoxia conditions again. This kind of conversion from normoxia to hypoxia lasted about 5 days. At 24, 48, 72, and 150 h, the shrimp were sampled, and their hemocytes and gills were collected. At the same time as the hypoxia experiment, six shrimp which were kept continuously in normoxia condition for 150 h were collected as control.
RNA extraction and reverse transcription
Total RNA was extracted from hemocytes, gill, muscle, intestine, ovary, lymphoid organ, hepatopancreas, or cephalothorax with TRIzol Reagent (Invitrogen, USA) as described in the manufacturer’s protocol. RNA quality was assessed by electrophoresis on 1% agarose gel.
For reverse transcription, the cDNA was synthesized in a 25-μl reaction volume containing 2 μg of DNase I-treated total RNA, 1× Moloney murine leukemia virus (MMLV) buffer, 0.5 mM deoxyribonucleotide triphosphate (dNTP), 0.4 mM oligo-dT, 20 U of RNase inhibitor (Promega), and 200 U of MMLV reverse transcriptase (Promega). The cDNA was then diluted five times, and 1 μl of the dilution was used for each RT-PCR reaction.
Cloning and sequencing of FcHSP90 cDNA fragment
The Chinese shrimp cephalothorax cDNA library was constructed by the ZAP express cDNA synthesis kit (Stratagene, USA) and ZAP expression cDNA Gigapack® III Gold cloning kit (Stratagene, USA) and was used to create templates for the following PCR amplifications. An expressed sequence tag (EST) of HSP90 was found in large-scale EST sequencing of shrimp cephalothorax cDNA library (Xiang 2002). The EST sequence was confirmed by amplification and sequencing using the specific primers, FcHSP90f1 and FcHSP90r1 (see Table 1).
Table 1.
Nucleotide sequences and positions of primers used in polymerase chain reaction (H = T/C/A, Y = T/C, R = A/G, K = T/G, N = A/T/G/C)
| Primer | Direction | sequence (5′ to 3′) | Position |
|---|---|---|---|
| FcHSP90f1 | F | AAGAACGACAAGTCAGTG | 2039 |
| FcHSP90r1 | R | TGCTGGTTTAAATCGTATG | 2430 |
| FcHSP90f2 | F | ACCTTCTACAGYAAYAARGAG | 182 |
| FcHSP90r2 | R | TTCYTCHARGTACTCNGTCTG | 676 |
| FcHSP90f3 | F | TCTCGCATGGAAGAAGTCG | 2255 |
| FcHSP90r3 | R | AGAAGCCCACGCCGAACTGA | 491 |
| FcHSP90f4 | F | GAACAACGACGACGAACAGT | 535 |
| FcHSP90r4 | R | TGAAGCCAGATGACAGAAGG | 2108 |
| FcHSP90f5 | F | GACCGCACGCTCACCATCAT | 332 |
| 18sf | F | AGTAGCCGCCCTGGTTGTAGAC | |
| 18sr | R | TTCTCCATGTCGTCCCAGT |
Random primer NNNNNN
Two degenerated primers, FcHSP90f2 and FcHSP90r2, were designed based on the multiple alignment of HSP90 homologous sequences of insects (Apis mellifera XM_393090, Aedes aegypti DQ440225, Drosophila melanogaster NM_078577, and Bombyx mori NM_001043372), of fish (Danio rerio BC063946), and of humans (Homo sapiens NM_005347 9). The PCR amplification was primed by FcHSP90f2 and FcHSP90r2, and the thermal profile consisted of an initial denaturation (94°C, 5 min), followed by 35 cycles of denaturation (94°C, 50 s), annealing (54°C, 50 s) and extension (72°C, 90 s), and a final extension step (72°C, 10 min). A gene-specific forward primer FcHSP90f4 was designed based on this PCR product. The reverse primer FcHSP90r4 was designed according to the EST sequence. The amplification by FcHSP90f4 and FcHSP90r4 was performed as following: an initial denaturation (94°C, 5 min), followed by 35 cycles of denaturation (94°C, 50 s), annealing (62°C, 50 s) and extension (72°C, 100s), and a final extension (72°C, 10 min) step.
The specific primer FcHSP90f3 was designed according to the EST and employed for the 3′ end amplification with the universal primer T7. A reverse gene specific primer FcHSP90r3 was designed based on the fragment amplified by the primers FcHSP90f2 and FcHSP90r2. With FcHSP90r3, the universal primer T3 was used for amplifying the 5′ end. PCR was performed using PTC-100 (MJ Research Company, USA) as following: an initial denaturation (94°C, 5 min), followed by 35 cycles of denaturation (94°C, 50 s), annealing (55°C, 50 s) and extension (72°C, 70 s), and a final extension step (72°C, 10 min).
HSP90 gene-specific primers FcHSP90f4 and FcHSP90r4 were designed based on the 5′ and 3′ sequences cloned above, respectively. The thermo profile was carried out as following: 94°C for 5 min, followed by 35 cycles of denaturation (94°C, 1 min), annealing (60°C, 1 min) and extension (72°C, 2 min 30 s), and a final extension step (72°C, 10 min). The sequences of all primers mentioned above are listed in Table 1. All primers were synthesized in Shenggong Biocompany (Shanghai, China).
All PCR reactions above were carried out in a 25 μl reaction system: 1U Takara Ex taq hot start, 1× Ex taq buffer (plus Mg2+), 0.2 mM dNTP mixture, 0.2 mM forward primer, 0.2 mM reversed primer and 1 μl cDNA template. PCR products were analyzed on 1.5% agarose gels, extracted with a QIA quick gel extraction kit (QIAGEN), and cloned into pMD18-T vector (TaKaRa). The resultant recombinant plasmid was then transformed into E. coli host TOP 10′. The positive transformers were screened by PCR and then sequenced in Shenggong Biocompany (Shanghai, China).
Analysis of FcHSP90 cDNA and deduced amino acid sequences
The FcHSP90 cDNA sequence was analyzed by BioEdit (version 7.0.1) software package, and the deduced amino acid sequence was predicted by the open reading frame (ORF) finder at the National Center for Biotechnology Information web-site (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The deduced amino acid sequence was analyzed by Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de). The Compute pI/Mw (http://www.expasy.ch/tools/pi_tool.html) was used to calculate the theoretical isoelectric point (pI) and molecular weight (Mw). The amino acid sequences of different species were obtained from the GenBank database (http://www.ncbi.nih.gov), and a multiple sequence alignment was created with ClustalX (version 1.83). Subsequently, the neighbor-joining phylogeny tree was generated by MEGA (version 3.1).
Quantitative analysis of HSP90 mRNA expression by real-time RT-PCR
A pair of gene-specific primers, FcHSP90f5 and FcHSP90r3, were designed to amplify a 125-bp product. A pair of 18s rRNA gene-specific forward and reverse primers, 18sf and 18sr (Table 1), were designed to amplify a 147-bp long fragment of 18s rRNA. Nuclease-free water (Promega, USA) was used instead of cDNA templates as PCR negative control.
A quantitative real-time RT-PCR method was carried out using Mastercycler ep realplex 4 (Eppendorf, Germany) to analyze the mRNA expression profiles of FcHSP90 in shrimp. The real-time RT-PCR reactions were carried out in 25 μl reaction systems with 1 U Takara Ex taq hot start, 1× Ex taq buffer (plus Mg2+), 0.2 mM dNTP mixture, 1× SYBR Green Master Mix (Applied Biosystems, USA), 0.2 mM forward primer, 0.2 mM reversed primer, and 1 μl cDNA template. The thermal profile for real-time RT-PCR was 95°C for 2 min, followed by 40 cycles of 95°C 15 s, 55°C 15 s, and 68°C 15 s. In order to confirm that only one specific PCR product was amplified, a melt cycle, in which PCR product was denatured from 68°C to 100°C, was added to each thermal profile to produce the melt curves. The fluorescent real-time PCR data were analyzed by RealPlex Software (Eppendorf, Germany). The baselines were set automatically by the software taking consistency into account. To analyze expression profile of FcHSP90, the comparative CT value method (Livak and Schmittgen 2001) was employed. SPSS 11.0 Software was employed to determine variations in the data obtained from fluorescent real-time PCR using one-way analysis of variance followed by the Tukey honestly significant difference test. Significance was concluded at P < 0.05.
Results
cDNA cloning and sequence analysis of FcHSP90
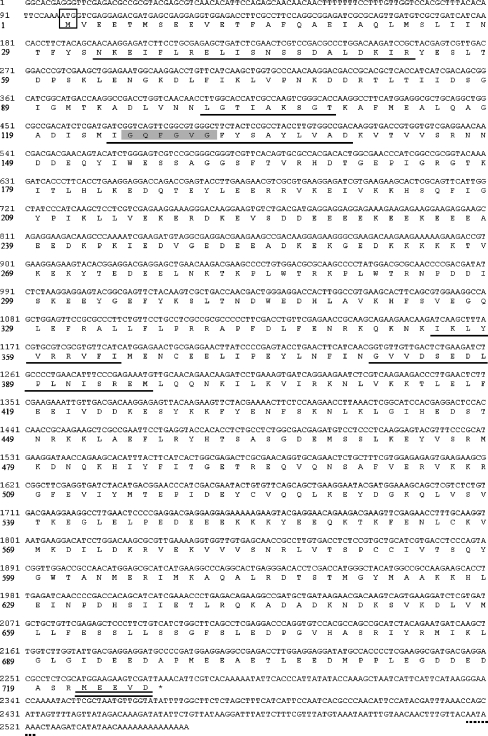
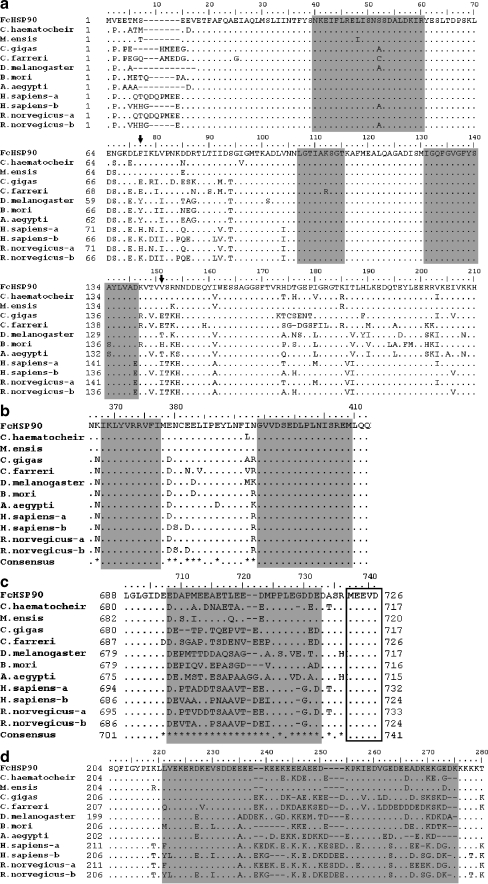
The full length of FcHSP90 cDNA sequence (Genbank accession no. EF032650) was obtained by PCR amplification. The full-length FcHSP90 is 2,552 bp and contains a 2,181-bp ORF encoding 726 amino acids (Fig. 1). 5′-untranslated region (UTR) is 97 bp, and 3′ UTR is 372 bp with a canonical polyadenylation signal sequence AATAAA and a poly (A) tail. The predicted Mw of FcHSP90 is 84.11 kDa, and theoretical pI is 4.93. Three conserved functional domains were found in FcHSP90: N domain (1–220), middle domain (M domain, 255–540), and C domain (541–726). Five conserved HSP90 protein family signatures were found in the deduced amino acid sequence of FcHSP90 (Fig. 1): NKEIFLRELISNSSDALDKIR (33–53), LGTIAKSGT (100–108), and IGQFGVGFYSAYLVAD (124–139) were located at N domain (Fig. 2a); IKLYVRRVFI (355–364) and GVVDSEDLPLNISREM (381–396) were located in the M domain (Fig. 2b). A typical histidine kinase-like ATPase domain is located at 33–187 of FcHSP90 using SMART analysis. The conserved “GxxGxG” motif was found in M domain (Fig. 2b). The C-terminal-conserved MEEVD motif was also found in the FcHSP90 sequence (Fig. 2c).
Fig. 1.
Complete full-length FcHSP90 cDNA and deduced amino acid sequences. Start codon (ATG) is boxed. Asterisk indicates stop codon (TAA). Five HSP90 protein family signatures are underlined. Conserved “GxxGxG”motif is shaded with gray background. Conserved “MEEVD” motif is double-underlined. The polyadenylation signal (AATAAA) is underlined by a dotted line
Fig. 2.
Homology analysis of FcHSP90 deduced amino acid sequence aligned by Bioedit 7.0. a The alignment of N domain of HSP90s. Three HSP90 protein family signatures are shaded with gray background. Arrows indicate phenylalanine (F, 77) and valine (V, 151) presenting homology divergence. b The alignment of M domain of HSP90s. Two HSP90 protein family signatures are shaded with gray background. c The alignment of C terminus of HSP90s. The residues shaded are variable parts located at the C terminus of HSP90s. The conserved “MEEVD” motif is boxed. d The link segment between N and M domain. The variable part is shaded with gray background. Residues identical to FcHSP90 at the same point in alignment are replaced by dot. Consensus sequence is produced: dots indicate at the point that all residues are identical and asterisks indicate that there are variations. Abbreviations and accession numbers are shown as follows: FcHSP90, F. chinensis HSP90, EF032650; C. haematocheir, C. haematocheir HSP90, AAS19788; M. ensis-HSP90, M. ensis HSP90, ABR66910; C. gigas, C. gigas HSP90,ABS18268; C. farreri, C. farreri AAR11781; D. melanogaster-HSP90, D. melanogaster HSP90, NP_523899; B. mori, B. mori HSP90, BAB41209; A. aegypti, A. aegypti HSP90, XP_001649752; H. sapiens-a and -b H. sapiens HSP90α, NP_005339, H. sapiens HSP90 β,NP_031381; R. norvegicus-a and –b, Rattus norvegicus HSP90α, NP_786937, R. norvegicus HSP90β, P34058)
Homology analysis of FcHSP90
The alignment of the HSP90s observed show that shrimp FcHSP90 protein has a high degree of sequence homology with HSP90s from other species (Fig. 2). FcHSP90 showed a much greater similarity (96% and 89%, respectively) to those of crustaceans Metapenaeus ensis and Chiromantes haematocheir compared with those of vertebrates and insects. Three functional domains (N, M, and C domain) were shown to be highly conserved (Fig. 2). The alignment result showed that the N and M domain segment links were variable among HSP90s of different species (Fig. 2d). About 20 residues at the head of the conserved “MEEVD” motif were shown to be variable (Fig. 2c). Several residues such as phenylalanine (F, 77) and Valine (V, 151) were found to be conserved inside the group but variable among groups (Fig. 2a).
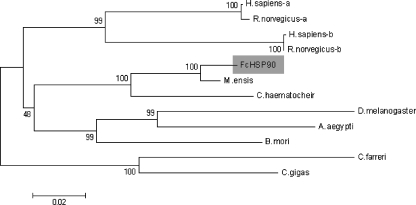
The neighbor-joining phylogeny tree was consistent with the alignment result. Three major groups clustered in the phylogeny tree: vertebrate, arthropod, and mollusk groups, while the crustacean and insect subgroups can be seen in the arthropod group. FcHSP90 was located in the crustacean subgroup and was observed to be closer to HSP90s of other crustaceans (Fig. 3).
Fig. 3.
The neighbor-joining tree shows the relationship of FcHSP90 with other known HSP90s. Alignment of amino acid sequences are produced by Clustal W, and the bootstrap neighbor-joining phylogeny tree is constructed by MEGA 3.1 (bootstrap = 100). FcHSP90 is shaded with gray background. Abbreviations and accession numbers are the same as shown in Fig. 2
Tissue distribution of FcHSP90 mRNA
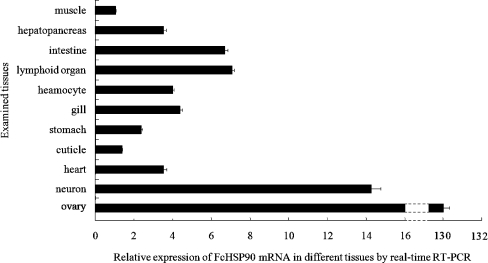
The mRNA transcripts of FcHSP90 were widely detected in all examined tissues at different expression levels (Fig. 4). The highest expression was observed in ovary, in which the FcHSP90 mRNA transcript level was found at levels 130-fold higher than that in muscle.
Fig. 4.
Quantitative analysis of FcHSP90 mRNA in different tissues of F. chinensis by real-time RT-PCR. The expression amount of FcHSP90 in muscle was taken as the base amount (=1). Error bars represent the ±SD of six shrimp across three independent PCR amplifications and quantifications (n = 6)
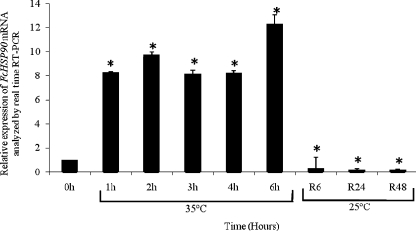
Expression profile of FcHSP90 under heat shock treatment
The relative expression level of FcHSP90 mRNA in juvenile shrimp is shown in Fig. 5. At the first hour during heat shock, the expression level of FcHSP90 mRNA in whole shrimp was significantly elevated (8.3-fold) compared to that at 0 h before heat shock treatment, and the shrimp maintained their high levels through 2, 3, and 4 h until reaching the highest level at 6 h during heat shock treatment, which was shown to be 12.3-fold higher than that of the control at the starting point. During the recovery period after shrimp were transferred from heat shock condition (35 ± 1°C) to normal condition (25 ± 1°C), the mRNA transcription levels of FcHSP90 dropped sharply to a level which was significantly lower than that of the control.
Fig. 5.
FcHSP90 relative expression in juvenile shrimp (F. chinensis) after heat shock treatment analyzed by real-time RT-PCR. Six individuals for each group at sample points: 0, 1, 2, 3, 4, and 6 h during heat shock at 35°C, as well as at 25°C for 6 h (R6), 24 h (R24) and 48 h (R48) of the recovery period. Asterisk indicates significant differences compared to that at the start point without heat shock (P < 0.05). Error bars represent the ±SD of six shrimp across three independent PCR amplifications and quantifications (n = 6)
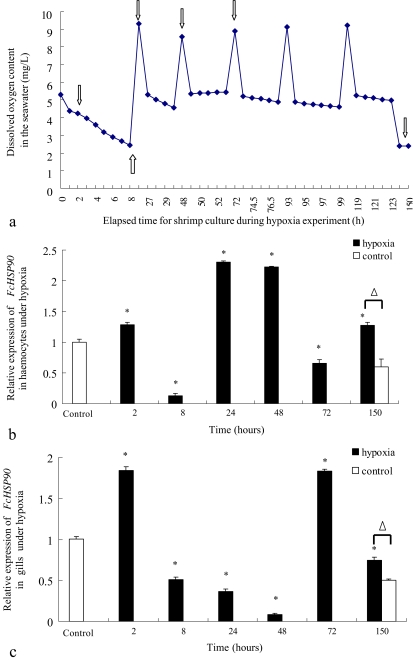
Expression profile of FcHSP90 mRNA under hypoxia conditions
The shrimp experienced a series of variations of O2 concentration in sea water for hypoxia experiments. The recorded O2 concentration during the whole experiment and the sampling point was shown in Fig. 6a. The time course analysis of FcHSP90 in hemocytes and gill of Chinese shrimp were analyzed using real-time RT-PCR. Distinct expression profiles of FcHSP90 in hemocytes (Fig. 6b) and gill (Fig. 6c) of F. chinensis were observed under hypoxia conditions.
Fig. 6.
Record of dissolved oxygen concentration of seawater in the cultured container during hypoxia experiment (a), the FcHSP90 relative expression profile in hemocytes of F. chinensis during hypoxia (b), and that in gills of F. chinensis during hypoxia (c) analyzed by real-time RT-PCR. The hollow arrows in a refer to the sampling points in the hypoxia experiment. Asterisk represents significant difference between the expression level of hypoxia-stressed shrimp at the sampling point and that of the control shrimp before the hypoxia experiment (P < 0.05). Open triangle indicates the significant differences between the expression level of shrimp under hypoxia conditions for 150 h and that at 150 h of control shrimp under normoxia conditions. Error bars represent the ±SD of six shrimp across three independent PCR amplifications and quantifications (n = 6)
At 2 h during hypoxia stress, the transcription levels of FcHSP90 in hemocytes and gills were up-regulated compared with that in shrimp without any hypoxia stress (P < 0.05). At 8 h during hypoxia stress, its transcription levels in hemocytes and gills were greatly down-regulated compared with that in the control shrimp. At 24, 48, and 72 h, when the O2 concentration was restored to the normoxia conditions from hypoxia conditions, the transcription levels of FcHSP90 in hemocytes were apparently up-regulated at 24 and 48 h but down-regulated at 72 h compared with that in the control shrimp. In contrast, the transcription level of FcHSP90 in the gills was still down-regulated at 24 and 48 h but up-regulated at 72 h. At 150 h, when the shrimp had suffered conversions from hypoxia to normoxia for 5 days and the O2 concentration of sea water was dropped to 28% O2 saturation, the transcription levels of FcHSP90 in hemocytes and gills were up-regulated compared with the shrimp cultured in normoxia condition all the time.
Discussion
HSP90, co-chaperoned with other proteins such as the HSP70 family proteins (Pratt and Toft 2003), is a highly conserved and abundant cytoplasmic protein involved in protein folding, cytoprotection, proteosomic degradation and a number of cellular regulatory pathways (Minami et al. 2000; Hartl and Hayer-Hart 2002; Zhang and Burrows 2004; Brown et al. 2007). In this study, the full-length cDNA sequence of cytoplasmic HSP90 (FcHSP90) was cloned from Chinese shrimp F. chinensis. Five conserved HSP90 family signatures were identified in the deduced FcHSP90 amino acid sequence. Conserved domains of FcHSP90 guarantee the conserved functions of HSP90 such as ATP hydrolysis and client protein binding (Hartl and Hayer-Hart 2002; Brown et al. 2007). In the C domain of FcHSP90, about 20 residues near the C terminus were highly variable among different species. The conserved “MEEVD” motif located at the C terminus of FcHSP90 indicated that FcHSP90 activities can probably be mediated by proteins containing tetraticopeptide repeat domains as in other species (McDonough and Patterson 2003; Brown et al. 2007). In the M domain, the conserved “GxxGxG” motif, which was predicted and which wraps around ATP in the tertiary structure, was found (Pearl and Prodromou 2006). The phylogeny tree revealed that FcHSP90 belonged to the invertebrate group and was located in the crustacean branch. Both the alignment result and the phylogeny tree analysis showed that FcHSP90 was more homologous with HSP90s of previously identified crustaceans. As shown by protein alignment (Fig. 2a), the phenylalanine (F, 77) is conserved in Crustacea, while the homologous amino acid at this position diverges as glutamic acid (E) in Mollusca (Crassostrea gigas and Chlamys farreri), tyrosine (Y) in insects (D. melanogaster, B. mori, and A. aegypti), lysine (K) in rat, and histidine (H) in humans; the valine (V, 151) in Crustacea corresponding to glutamic acid (E) in Mollusca, threonine (T) in Diptera, histidine (H) in Lepidoptera, and isoleucine (I) in rat and humans. This sort of homology divergence possibly qualifies HSP90 as a potential evolutionary marker at a particular evolutionary stage of life (Gupta 1995). The variable residues such as phenylalanine (F, 77) and valine (V, 151) in FcHSP90 would possibly qualify HSP90 as a potential molecular marker in every stage of eukaryotic organisms (Gupta 1995).
The real-time RT-PCR analysis showed that FcHSP90 was ubiquitously expressed in the examined tissues of F. chinensis. The highest expression was detected in ovary of shrimp. Previous reports demonstrated that HSP90 was essential to phosphorylation of Mos, a proto-oncogene protein, and to activation of the mitogen-activated protein kinase pathway that is responsible for oocyte maturation in Xenopus (Ali et al. 1998; Fisher et al. 2000). Therefore, we speculate that the extraordinary expression detected in ovary suggests that FcHSP90 might mediate a similar cascade regulating ovary maturation in F. chinensis.
When we studied the gene expression of FcHSP90 in response to heat shock, we used the juveniles to do the heat shock experiments. During the culture of F. chinensis in shrimp pond, the natural high temperature usually occurs at early August, and the shrimp size is not large at this time. So we chose juvenile to do heat shock experiment in order to make it more close to the actual situation. We did not separate different tissues since the hemolymph is difficult to isolate at this stage. The whole cephalothoraxs of the juveniles were used to extract RNA, so the detected expression can reflect the actual situation of the whole shrimp.
The expression of FcHSP90 was very sensitive to heat shock. At the first hour post heat shock, the mRNA level of FcHSP90 increased significantly and rapidly and reached levels 8.3-fold higher than that at the start time. The highest expression level was detected at 6 h during heat shock. The data suggested that FcHSP90 might be directly involved in the resistance of organisms to heat shock. It was reported that HSP90 was likely involved in heat shock transcription factor trimerization to activate a target gene and to protect the cell against death after heat shock (Ali et al. 1998). HSP90 could also protect the cell against heat shock-induced apoptosis (Malago et al. 2002; Spees et al. 2002; Yavelsky et al. 2004). It was also reported that the transcription of HSP90 could be induced by heat shock in other aquatic animals such as the abalone H. tuberculata (Farcy et al. 2007) and the Zhikong scallop C. farreri (Gao et al. 2007).
However, mRNA levels of FcHSP90 during heat shock recovery were significantly lower than that at 0 h. When the shrimp culture temperature changed from high temperature (35°C) to low temperature (25°C), a cold shock might have inhibited the transcription of FcHSP90. A similar expression profile of HSP90 was also reported in the American lobster Homarus americanus during heat shock recovery (Malago et al. 2002).
Hypoxia is a very common stress factor encountered in shrimp culture ponds. At night, the shrimp are more likely to stay at the bottom of the pond, where hypoxia conditions are more likely to occur. We therefore mimic the hypoxia conditions in shrimp ponds when we designed the experiment. Since hypoxia usually occurs at the late phase of culture when the shrimp grows to mature stage, mature shrimp were chosen to study the expression of FcHSP90 in different tissues. To different genes, their expression profiles in different tissues are usually different, so we chose different tissues to study the gene expression.
When shrimp were transferred from normoxia to hypoxia for a short period (2 h), the transcription of FcHSP90 was apparently elevated due to hypoxia. If the hypoxia condition lasts for a longer time (8 h), the transcription of FcHSP90 will be greatly inhibited. When the sea water was restored to 100% O2 saturation followed by a long time of hypoxia, the expression of FcHSP90 in the hemocytes was greatly elevated, but it was still kept at very low levels in the gills. After 5 days of hypoxia stress, the FcHSP90 expression would still be up-regulated compared with that of the controls at the same time. The data showed that hypoxia caused the large variation of transcription of FcHSP90. After an extended period of hypoxia, the expression of FcHSP90 in different tissues was changed greatly even if the culture conditions were restored to normoxia. We inferred that hypoxia can change the expression level of FcHSP90 significantly. Comparing the expression profiles of FcHSP90 between hemocytes and gills, we inferred that gills need more time to adjust the gene expression of FcHSP90 after a long period of hypoxia. This difference might depend on the different roles that these two tissues play during hypoxia stress. Similar results have been reported in C. gigas that hypoxia causes the variation in levels of heat shock proteins including HSP70 and HSP25, and different expression profiles were obtained for HSP70 in different tissues after hypoxia stress (David et al. 2005). HSP90 can modulate the stabilization of hypoxia-inducible factor-1α (HIF-1α) and subsequently change the conformation of the heterodimeric complex comprised of HIF-1α and aryl hydrocarbon receptor nuclear translocator to regulate the gene expression for cytoprotection under hypoxia (Ali et al. 1998; Hur et al. 2002; Isaacs et al. 2002, 2004).
It is worth pointing out that the transcription level of FcHSP90 in shrimp which were kept in normoxia sea water for 150 h (about 6 days) declined compared with that at 0 h control. It means that there are other factors which can affect the transcription level of FcHSP90 in addition to hypoxia. Therefore, we suggest that a parallel control experiment should be set if the hypoxia experiment lasts more than 3 days. Since the culture conditions are difficult to keep stable, some other conditions such as confinement may affect gene expression. Suitable parallel control experiments can help to exclude the effect of other factors on gene expression. In a previous report of hypoxia experiments in C. gigas, the expression levels of several genes including carbonic anhydrase, glutathione peroxidase, metallothionein, HSP70, etc. varied greatly in the control group after 3 days (David et al. 2005). Due to the sensitivity of FcHSP90 expression in the shrimp response to heat shock and hypoxia, FcHSP90 might play very important roles to allow shrimp to cope with environmental stresses.
Acknowledgements
This work was supported by the National High-Tech Research and Development Program of China (863 program) 2006AA09Z424 and 2006AA10A402, the Major State Basic Research Development Program of China (973 program) 2006CB101804.
References
- Ali A, Bharadwaj S, O’Carroll R, Ovsenek N. HSP90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes. Mol Cell Biol. 1998;18:4949–4960. doi: 10.1128/mcb.18.9.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachère E, Chagot D, Grizel H. Separation of Crassostrea gigas hemocytes by density gradient centrifugation and counterflow centrifugal elutriation. Dev Comp Immunol. 1988;12:549–559. doi: 10.1016/0145-305X(88)90071-7. [DOI] [PubMed] [Google Scholar]
- Bendena WG, Southgate AA, Garbe JC, Pardue ML. Expression of heat shock locus hsr-omega in nonstressed cells during development in Drosophila melanogaster. Dev Biol. 1991;144:65–77. doi: 10.1016/0012-1606(91)90479-M. [DOI] [PubMed] [Google Scholar]
- Brown MA, Zhu L, Schmidt C, Tucker PW. Hsp90-From signal transduction to cell transformation. Biochem Biophys Res Commun. 2007;363:241–246. doi: 10.1016/j.bbrc.2007.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellura C, Toubiana M, Parrinello N, Roch P. HSP70 gene expression in Mytilus galloprovincialis hemocytes is triggered by moderate heat shock and Vibrio anguillarum, but not by V. splendidus or Micrococcus lysodeikticus. Dev Comp Immunol. 2006;30:984–997. doi: 10.1016/j.dci.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Currie S, Tufts B. Synthesis of stress protein 70 (Hsp 70) in rainbow trout (Oncorhynchus mykiss) red blood cells. J Exp Biol. 1997;200:607–614. doi: 10.1242/jeb.200.3.607. [DOI] [PubMed] [Google Scholar]
- David E, Tanguy A, Pichavant K, Moraga D. Response of the Pacific oyster Crassostrea gigas to hypoxia exposure under experimental conditions. FEBS J. 2005;272:5635–5652. doi: 10.1111/j.1742-4658.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- Deane EE, Kelly SP, Luk JCY, Woo NYS. Chronic salinity adaptation modulates hepatic heat shock protein and insulin-like growth factor I expression in black sea bream. Mar Biotechnol. 2002;4:193–205. doi: 10.1007/pl00021690. [DOI] [PubMed] [Google Scholar]
- Farcy E, Serpentini A, Fiévet B, Lebel JM. Identification of cDNAs encoding HSP70 and HSP90 in the abalone Haliotis tuberculata: Transcriptional induction in response to thermal stress in hemocyte primary culture. Comp Biochem Physiol Part B. 2007;146:540–550. doi: 10.1016/j.cbpb.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Fisher DL, Mandart E, Doree M. Hsp90 is required for c-Mos activation and biphasic MAP kinase activation in Xenopus oocytes. EMBO J. 2000;19:1516–1524. doi: 10.1093/emboj/19.7.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Song LS, Ni DJ, Wu LT, Zhang H, Chang YQ. cDNA cloning and mRNA expression of heat shock protein 90 gene in the haemocytes of Zhikong scallop Chlamys farreri. Comp Biochem Physiol Part B. 2007;147:704–715. doi: 10.1016/j.cbpb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Phylogenetic analysis of the 90 kD heat shock family of protein sequences and an examination of the relationship among animals, plants, and fungi species. Mol Biol Evol. 1995;12:1063–1073. doi: 10.1093/oxfordjournals.molbev.a040281. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hart M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hur E, Kim HH, Choi SM, et al. Reduction of hypoxia-induced transcription through the repression of hypoxia-inducible factor-1α/aryl hydrocarbon receptor nuclear translocator DNA binding by the 90-kDa heat-shock protein inhibitor. Radicicol Mol Pharmacol. 2002;62:975–982. doi: 10.1124/mol.62.5.975. [DOI] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mimnaugh EG, Martinez A, Cuttitta F, Neckers LM. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J Biol Chem. 2002;277:29936–29944. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Neckers L. Aryl hydrocarbon nuclear translocator (ARNT) promotes oxygen-independent stabilization of hypoxia-inducible factor-1 by modulating an hsp90-dependent regulatory pathway. J Biol Chem. 2004;279:16128–16135. doi: 10.1074/jbc.M313342200. [DOI] [PubMed] [Google Scholar]
- Jiao CZ, Wang ZZ, Li FH, Zhang CS, Xiang JH. Cloning, sequencing and expression analysis of cDNA encoding a constitutive heat shock protein 70 (HSC70) in Fenneropenaeus chinensis. Chi Sci Bull. 2004;49:2385–2393. doi: 10.1360/982004-120. [DOI] [Google Scholar]
- Kültz D. Plasticity and stressor specificity of osmotic and heat shock responses of Gillichthys mirabilis gill cells. Am J Physiol. 1996;271:C1181–C1193. doi: 10.1152/ajpcell.1996.271.4.C1181. [DOI] [PubMed] [Google Scholar]
- Lang L, Miskovic D, Lo M, Heikkila JJ. Stress-induced, tissue specific enrichment of hsp 70 mRNA accumulation in Xenopus laevis embryos. Cell Stress Chaperones. 2000;5:36–44. doi: 10.1379/1466-1268(2000)005<0036:SITSEO>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yang WJ, Zhu XJ, Karouna-Renier NK, Rao RK. Molecular cloning and expression of two HSP70 genes in the prawn, Macrobrachium rosenbergii. Cell Stress Chaperones. 2004;9:313–323. doi: 10.1379/CSC-40R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the
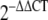 Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar] - Lo WY, Liu KF, Liao IC, Song YL. Cloning and molecular characterization of heat shock cognate 70 from tiger shrimp (Penaeus monodon) Cell Stress Chaperones. 2004;9:332–343. doi: 10.1379/CSC-47R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malago JJ, Koninkx JF, Dijk JE. The heat shock response and cytoprotection of the intestinal epithelium. Cell Stress Chaperones. 2002;7:191–199. doi: 10.1379/1466-1268(2002)007<0191:THSRAC>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:CALBTC>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y, Kawasaki H, Minami M, Tanahashi N, Tanaka K, Yahara I. A critical role for the proteasome activator PA28 in the Hsp90-denpendent protein refolding. J Biol Chem. 2000;275:9055–9061. doi: 10.1074/jbc.275.12.9055. [DOI] [PubMed] [Google Scholar]
- Molina A, Biemar F, Müller F, Iyengar A, Prunet P, Maclean N, Martial JA, Muller M. Cloning and expression analysis of an inducible HSP70 gene from tilapia fish. FEBS Letters. 2000;474:5–10. doi: 10.1016/S0014-5793(00)01538-6. [DOI] [PubMed] [Google Scholar]
- Muller L, Schaupp A, Walerych D, Wegele H, Buchner J. Hsp90 regulates the activity of wild type p53 under physiological and elevated temperatures. J Biol Chem. 2004;279:48846–48854. doi: 10.1074/jbc.M407687200. [DOI] [PubMed] [Google Scholar]
- Myrmel T, McCully JD, Malkin L, Krukenkamp IB, Levitsky S. Heat shock protein 70 mRNA is induced by anaerobic metabolism in rat hearts. Circulation. 1994;90:299–305. [PubMed] [Google Scholar]
- Palmisano AN, Winton JR, Dickhoff WW. Tissue-specific induction of Hsp90 mRNA and plasma cortisol response in chinook salmon following heat shock, seawater challenge, and handling challenge. Mar Biotechnol. 2000;2:329–338. doi: 10.1007/s101260000005. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Piano A, Franzellitti S, Tinti F, Fabbri E. Sequencing and expression pattern of inducible heat shock gene products in the European flat oyster, Ostrea edulis. Gene. 2005;361:119–126. doi: 10.1016/j.gene.2005.06.034. [DOI] [PubMed] [Google Scholar]
- Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Prodromou C, Roe SM, Piper PW, Pearl LH. A molecular clamp in the crystal structure of the N-terminal domain of the yeast Hsp90 chaperone. Nat Struct Biol. 1997;4:477–482. doi: 10.1038/nsb0697-477. [DOI] [PubMed] [Google Scholar]
- Sanders BM, Martin LS, Nelson WG, Phelps DK, Welch W. Relationship between accumulation of a 60 kDa stress protein and scope for growth in Mytilus edulis exposed to a range of copper concentrations. Mar Environ Res. 1991;31:81–97. doi: 10.1016/0141-1136(91)90021-Y. [DOI] [Google Scholar]
- Schill RO, Görlitz H, Köhler HR. Laboratory simulation of a mining accident: acute toxicity, hsc/hsp70 response, and recovery from stress in Gammarus fossarum (Crustacea, Amphipoda) exposed to a pulse of cadmium. BioMetals. 2003;16:391–401. doi: 10.1023/A:1022534326034. [DOI] [PubMed] [Google Scholar]
- Schlesinger MJ. Heat shock proteins: a mini review. J Biol Chem. 1990;265:12111–12114. [PubMed] [Google Scholar]
- Song LS, Wu LT, Ni DJ, Chang YQ, Xu W, Xing KZ. The cDNA cloning and mRNA expression of heat shock protein 70 gene in the haemocytes of bay scallop (Argopecten irradians, Lamarck 1819) responding to bacteria challenge and naphthalin stress. Fish Shellfish Immunol. 2006;21:335–345. doi: 10.1016/j.fsi.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Spees JL, Chang SA, Snyder MJ, Chang ES. Thermal acclimation and stress in the American lobster, Homarus americanus: equivalent temperature shifts elicit unique gene expression patterns for molecular chaperones and polyubiquitin. Cell Stress Chaperones. 2002;7:97–106. doi: 10.1379/1466-1268(2002)007<0097:TAASIT>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa K, Minami M, Minam Y. Constantly updated knowledge of Hsp90. J Biochem. 2005;137:443–447. doi: 10.1093/jb/mvi056. [DOI] [PubMed] [Google Scholar]
- Travers SA, Fares MA. Functional coevolutionary networks of the Hsp70-Hop-Hsp90 system revealed through computational analyses. Mol Biol Evol. 2007;24:1032–1044. doi: 10.1093/molbev/msm022. [DOI] [PubMed] [Google Scholar]
- Williams JH, Farag AM, Stansbury MA, Young PA, Bergman HL, Petersen NS. Accumulation of HSP 70 in juvenile and adult rainbow trout gill exposed to metal-contaminated water and/or diet. Environ Toxicol Chem. 1996;15:1324–1328. doi: 10.1897/1551-5028(1996)015<1324:AOHIJA>2.3.CO;2. [DOI] [Google Scholar]
- Xiang JH (2002) Over 10, 000 expressed sequence tags from Fenneropenaeus chinensis. In: Abstract of International Aquaculture Conference and Exposition, Beijing, p 837
- Yavelsky V, Vais O, Piura B, Wolfson M, Rabinovich A, Fraifeld V. The role of Hsp90 in cell response to hyperthermia. J Therm Biol. 2004;29:509–514. doi: 10.1016/j.jtherbio.2004.08.078. [DOI] [Google Scholar]
- Zhang H, Burrows F. Targeting multiple signal transduction pathways through inhibition of Hsp90. J Mol Med. 2004;82:488–499. doi: 10.1007/s00109-004-0549-9. [DOI] [PubMed] [Google Scholar]