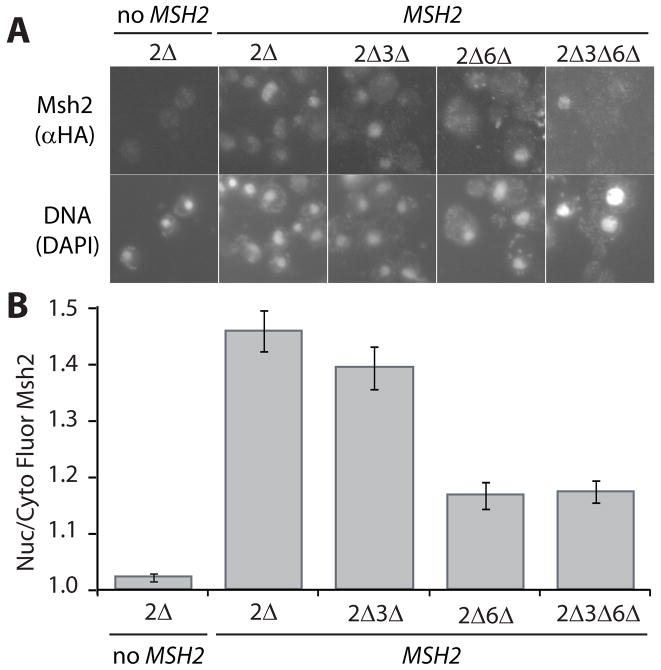
Fig. 1.
Indirect Immunofluorescence of Msh2 in the presence and absence of heterodimer partners Msh3 and Msh6. Yeast deletion strains (see Materials and Methods) lacking MSH2 (2Δ), MSH2 and MSH3 (2Δ3Δ), MSH2 and MSH6 (2Δ6Δ), and MSH2, MSH3 and MSH6 (2Δ3Δ6Δ) were transformed with a plasmid, pMSH2, expressing a hemagluttinin (HA) epitope-tagged Msh2 (MSH2). Cells lacking MSH2 transformed with a plasmid vector served as a negative control for background fluorescence (2Δ, no MSH2). Exponentially growing cells were prepared for immunofluorescence, incubated with mouse anti-hemaglutinin (α-HA) monoclonal primary antibody and goat α-mouse IgG Alexa Fluor 488 secondary antibody. (A) Representative Images of Msh2 in the presence and absence of heterodimer partners. Top panels are of Msh2 localization. Bottom panels show the nuclear staining of the cells in the top panels with the DNA specific fluorescent dye DAPI. (B) Quantitative Measurements of Nuclear Msh2. The nuclear and cytoplasmic fluorescence of Msh2 were determined using the ImageJ public domain Java image processing program [36]. The ratio of nuclear/cytoplasmic fluorescence (Nuc/Cyto Fluor Msh2) is plotted for each strain. The error bars signify the standard error of the mean. Approximately 50 cells per sample were analyzed.