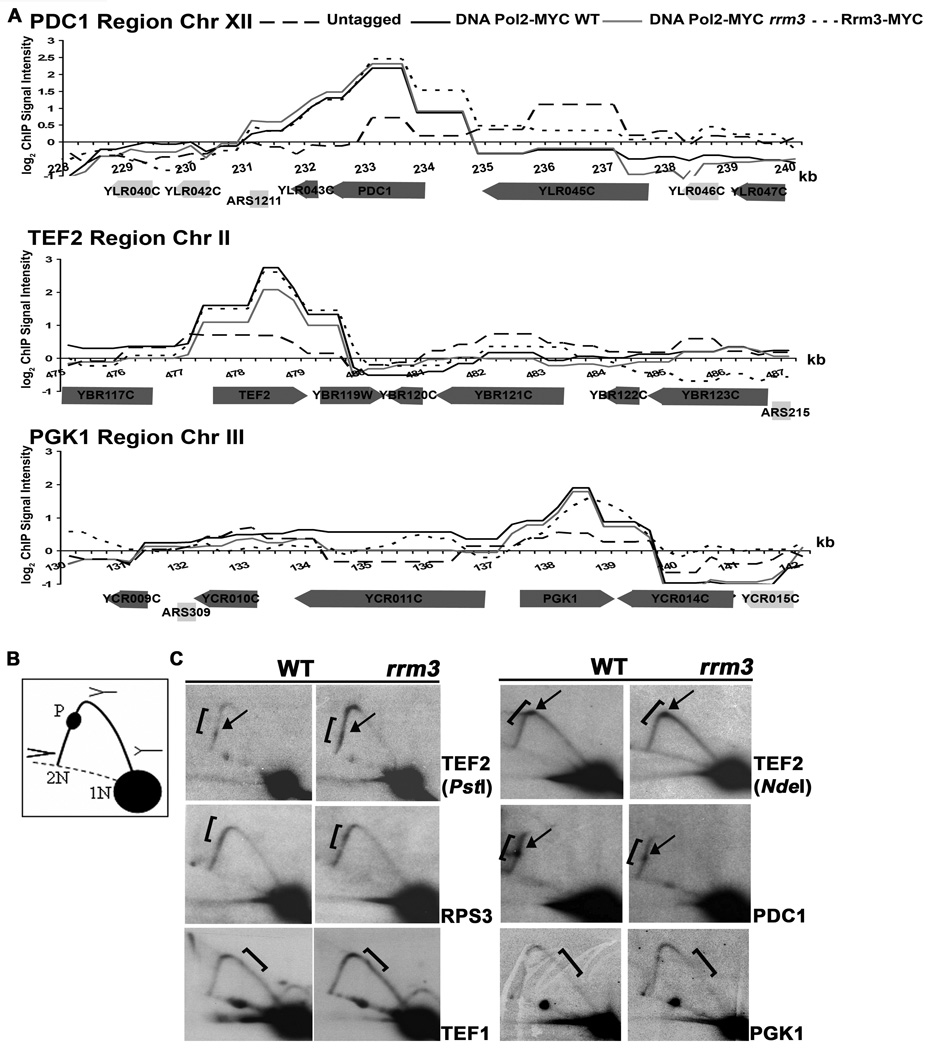
Figure 4. Highly transcribed genes are high occupancy DNA Pol2-MYC sites in WT and rrm3 cells as well as high occupancy Rrm3-MYC sites.
A. Association of DNA Pol2-MYC in WT and rrm3 cells and Rrm3-MYC with 12 kb regions on chromosomes XII, II and III, as in Fig. 1B. These regions contain the highly transcribed PDC1, TEF2, and PGK1 genes. Each gene was significantly associated with DNA Pol2-MYC in WT and rrm3 cells and with Rrm3-MYC. B. Schematics of restriction enzyme digested DNA with replication intermediates as visualized by 2D gels: 1N, non-replicating fragment; 2N, almost fully replicated fragment; P, replication pause; dotted line, arc of linear DNA molecules. C. DNA from WT and rrm3 cells was restriction enzyme digested, separated on 2D gels, and analyzed by Southern blotting at the indicated gene (restriction enzymes: EcoRV, TEF1 and PGK1; PstI, TEF2 and PDC1; BglII, RPS3; NdeI, TEF2). Brackets mark ORFs; arrows mark pauses. Pausing within TEF2 was also detectable in NdeI digested DNA when the pause was ~80% through the arc of replication intermediates.