Abstract
Cell adhesion to the extracellular matrix is mediated by elaborate networks of multiprotein complexes consisting of adhesion receptors, cytoskeletal components, signaling molecules, and diverse adaptor proteins. To explore how specific molecular pathways function in the assembly of focal adhesions (FAs), we performed a high-throughput, high-resolution, microscopy-based screen. We used small interfering RNAs (siRNAs) to target human kinases, phosphatases, and migration- and adhesion-related genes. Multiparametric image analysis of control and of siRNA-treated cells revealed major correlations between distinct morphological FA features. Clustering analysis identified different gene families whose perturbation induced similar effects, some of which uncoupled the interfeature correlations. Based on these findings, we propose a model for the molecular hierarchy of FA formation, and tested its validity by dynamic analysis of FA formation and turnover. This study provides a comprehensive information resource on the molecular regulation of multiple cell adhesion features, and sheds light on signaling mechanisms regulating the formation of integrin adhesions.
Introduction
Cell adhesion to the ECM is mediated via adhesion receptors, mainly integrins (Hynes, 1992), which are involved directly or indirectly in multiple processes including cell migration, morphogenesis, differentiation, and survival. When cells bind to an external surface, transmembrane multiprotein complexes such as focal adhesions (FAs) are formed, which consist of diverse scaffolding and signaling molecules (Geiger et al., 2001; Berrier and Yamada, 2007; Campbell, 2008). To date, >150 molecules, collectively known as the “adhesome” (Zaidel-Bar et al., 2007a; see http://www.adhesome.org), have been shown to reside, constitutively or transiently, in FAs and related integrin-mediated contacts (Geiger et al., 2001). These adhesion structures are highly dynamic, undergoing continuous assembly and disassembly during cell attachment and migration (Sastry and Burridge, 2000; Zamir et al., 2000; Kaverina et al., 2002).
Understanding this multicomponent and multifunctional system constitutes a major experimental challenge for researchers interested in structure–function relationships at adhesion sites.
A novel and powerful approach for addressing this challenge involves the application of the siRNA technique (Echeverri and Perrimon, 2006), which enables the specific perturbation of expression of selected genes. In this paper, we screened three siRNA libraries targeting genes encoding protein and lipid kinases and phosphatases, as well as a library targeting many of the known or suspected migration- and adhesion-related (MAR) genes (Simpson et al., 2008). High-resolution light microscopy, together with quantitative image analysis, was used to assess the effects of this treatment on the morphology of FAs, on their subcellular distribution, and on cell spreading and elongation (Liron et al., 2006; Paran et al., 2006).
To analyze the results of the screen, we took a systems biology approach, creating a multiparametric dataset of all the siRNAs that were found to induce significant changes (absolute z score > 3.5) in at least one of the FA or cell morphology features measured. This approach enabled us to analyze multiple effects with diverse strength. Analysis of these data revealed a high correlation between different FA morphological features (area, paxillin intensity, and length) in control and in most of the siRNA-treated cells.
Based on these correlations, we proposed a model for the hierarchical regulation of FA assembly. Informatic analysis yielded clusters of siRNAs, each of which induced a distinct “phenotypic signature.” Many of these clusters were enriched in siRNAs targeting genes involved in similar biological functions. Our screen sheds light on several principles of FA regulation, and highlights the involvement of specific genes in the orchestrated regulation of cell adhesion and morphogenesis.
Results
Screening for siRNAs affecting FAs and cell shape
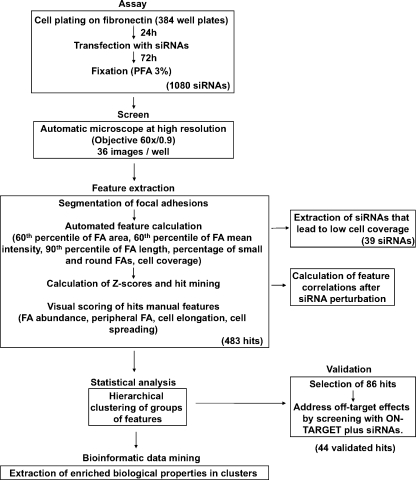
To identify genes involved in the regulation of FAs and cell shape, we conducted an siRNA screen using an automated, high-resolution, microscope-based assay (Fig. 1). For this purpose, an FA reporter cell line (HeLa cells expressing YFP-paxillin), specifically selected for its uniform FA morphology and distribution, was prepared (see Materials and methods). For the screen, cells were seeded on fibronectin-coated 384-well plates and transfected (24 h later) with the three human siRNA libraries chosen for this purpose: one targeting kinases (n = 576), a second targeting phosphatases (n = 192), and a custom library targeting MAR genes (n = 312). After fixation (72 h after transfection), wells were screened microscopically using a 60×/0.9 NA objective (Liron et al., 2006; Paran et al., 2006). A full screen of the three human siRNA libraries was repeated twice in duplicates, using SMARTpool concentrations of 100 nM and 50 nM.
Figure 1.
Workflow of the screen. Schematic description of the screen, including the cellular assay, automatic image acquisition, scoring of the images, statistical evaluation of the data (including data validation), and bioinformatics analysis.
Only plates with transfection efficiency better than 90%, based on the siTOX transfection control (which induces cell death; see Materials and methods), were taken for analysis. Moreover, we verified that the expression of fluorescent paxillin was suppressed by at least 99% using paxillin siRNA (Fig. S1).
The phenotypic features examined in this screen were subdivided into four main categories: cell coverage (percentage of total image area), which reflects cell number and spreading; FA morphology, which includes FA area (60th percentile, namely the FA area below which 60% of all FAs are found), FA mean intensity (60th percentile), FA length (90th percentile), and the percentage of small and round (dot-like) FAs, a rather prominent FA morphology feature; FA distribution, which includes FA abundance and peripheral FA; and cell shape, which includes spreading and elongation.
In this analysis, we define FAs as all distinct paxillin-containing structures larger than 0.25 µm2. Such structures correspond to bona fide FAs, as well as to focal complexes and fibrillar adhesions. The distinction between these structures is beyond the capacity of a single molecule (paxillin)-based high-throughput assay.
After the collection of images from all wells (36 high-resolution images per well), each FA morphology parameter was quantified and statistically evaluated. To identify siRNA hits, z scores (subtraction of the controls mean from each individual raw score, followed by division of the difference by the controls standard deviation) were calculated for each individual feature of each siRNA, based on comparison with the corresponding values of the control wells in the same plate.
Those siRNA phenotypes that displayed an absolute z score value of >3.5 in both screens (using 100 nM or 50 nM siRNA) for at least one parameter were considered hits (see the “Data analysis” in the Materials and methods section for a detailed description of the data analysis process, and for the assessment of experimental variability).
There was usually good agreement between the 100 nM and 50 nM screens, except that at 100 nM, many of the siRNAs induced lower cell coverage, and the percentage of toxic wells was high (21% compared with 3.6% at 50 nM). We therefore repeated the screen at 50 nM in order to lower the toxicity and off-target effects. siRNAs that were hits at 50 nM and toxic at 100 nM were included in the hit list.
For hit siRNAs, FA distribution features and cell shape features were scored manually, and compared with controls. Fig. S2 shows the reproducibility of each of the measured features; moreover, to assess the reproducibility of our measurements, we calculated the mean coefficient of variance (standard deviation divided by the mean of all the z scores) for each siRNA in the screen (displayed in Table S2).
We also visually examined all the screen images in order to exclude wells with highly heterogeneous cell culture or other technical artifacts. In this manner, we identified hits that in the visual scoring were significantly different from controls, but had no effect on FA morphology.
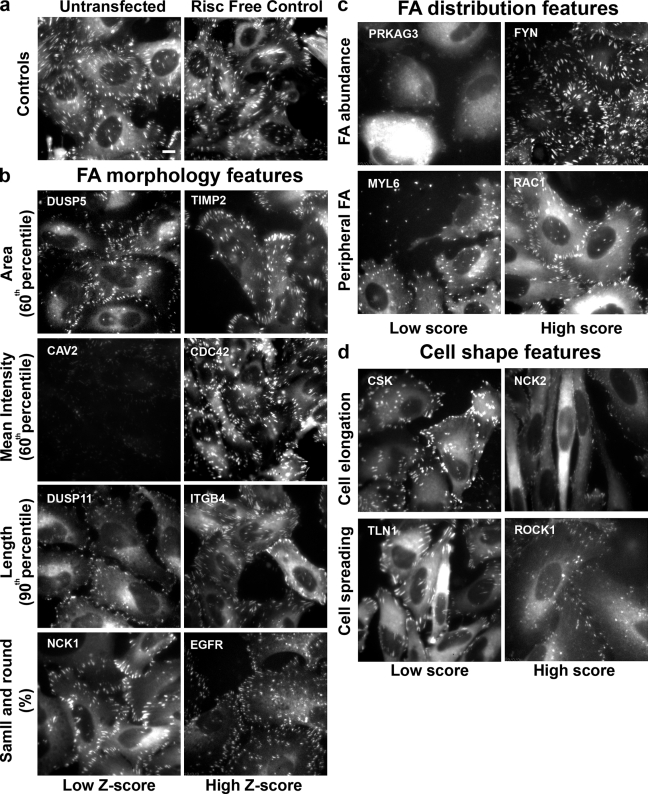
Fig. 2 depicts single images of sample wells in which treatment with the specified siRNAs led to phenotypes with a low or high z score or manual score in the analysis. The images are grouped according to the features analyzed: control cells (Fig. 2 a), FA morphological features (Fig. 2 b), FA distribution features (Fig. 2 c), and cell shape features (Fig. 2 d).
Figure 2.
Summary of the effects of siRNAs on the analyzed features. Single images of representative sample wells, showing significantly high or low z scores in FA morphological features or significantly high or low manual scores in FA distribution or cell shape features. The images in the “Low Z-score” or “Low score” columns are those with significantly low z scores or manual scores, respectively. The images in the “High Z-score” or “High score” columns are those with significantly high z scores or manual scores, respectively. (a) Untransfected and Risc-free–transfected HeLa cells expressing YFP-paxillin (controls). (b) Examples of hits that varied widely from control cells in their FA morphological features: FA area, FA intensity, FA length, and small and round FAs. (c) Examples of hits that varied significantly from control cells in FA distribution features: FA abundance, and peripheral FA. (d) Examples of hits that varied significantly from control cells in cell shape features: cell elongation and cell spreading. The siRNAs are designated by their Entrez gene symbol (National Center for Biotechnology Information). For full information concerning the hits, see Table S2. Bar (applies to all images), 10 µm.
Wells in which cell coverage was lower than 12% (altogether, 39 siRNAs in the 50-nM screen) were not scored for FA and cell shape features. Whether these siRNAs primarily target cell viability or cell adhesion remains to be explored. Nevertheless, based on the prominence, within this group, of siRNAs targeting cell cycle regulators and nuclear factor κB activation pathway components (Table S1), we assumed that most of the siRNAs affected cell number, target cell proliferation, and survival, rather than cell adhesion. The complete screening records, divided into hits, non-hits, and low cell coverage siRNA phenotypes, are available in Table S2.
The number of hits scoring positive for at least one parameter was very high, close to 45% of all siRNAs tested. The percentage of hits was particularly high among the MARs and phosphatases (53%), and lower among kinases (38%). The high frequency of hits may be attributed to the extensive involvement of these particular protein families in cell adhesion. Indeed, for individual features, the values were considerably smaller, ranging from 7.8% for cell elongation to 23.5% for FA length.
Analysis of the siRNA screen data
Cross-correlation between different FA morphological features in control cells.
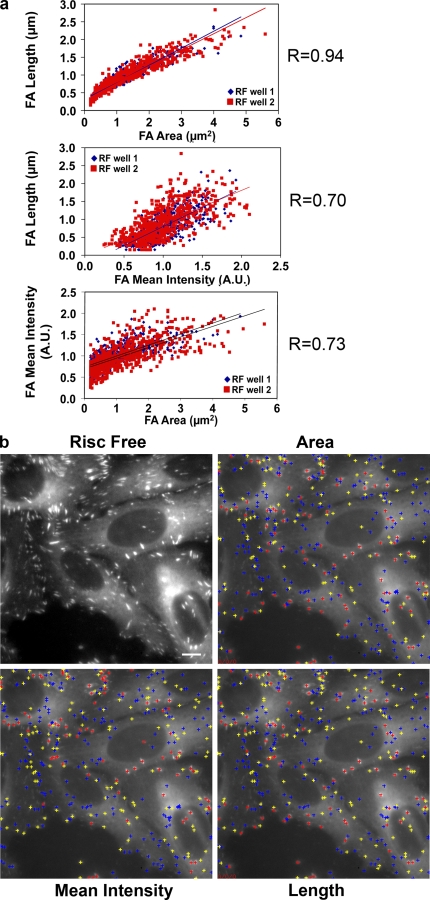
One advantage offered by a multiparametric screen is that it enables exploration of the interrelationships between the different structural features tested, irrespective of the particular siRNA treatment used. In control cells, we specifically calculated the correlations between FA area, mean intensity, and length. In Fig. 3 a, scatter plots of two control wells are presented, with each point representing a single FA. The data revealed a high correlation between FA area and length, suggesting that the growth of FAs is primarily attributable to elongation (R = 0.94). A high correlation between FA area and FA intensity was also observed, indicating that larger FAs (in area, R = 0.73; and in length, R = 0.70) also contain higher local concentrations of plaque proteins such as paxillin.
Figure 3.
Correlations between FA morphological features in control cells. (a) Scatter plots of FA morphological parameters from two different control wells (one high-resolution image per well) showing the correlation between the following FA morphological parameters: FA area, FA mean intensity (A.U., arbitrary units), and FA length. The mean correlation value (R) is shown. (b) A typical image of control cells (RISC Free), segmented in order to define FA objects and calculate their parameters. The centers of mass of the FAs are marked by crosses, color-coded according to the magnitude of the designated feature (area, mean intensity, and length). Red, high; yellow, medium; blue, low. Bar, 10 µm.
For a visual representation of these relationships, we marked the centers of mass of the FAs by crosses, color-coded according to the magnitude of the designated feature (area, mean intensity, or length). We found that the distribution of the colored crosses was closely similar between the images, which further indicates their correspondence to the correlation values (Fig. 3 b).
Clustering of siRNAs, based on their differential effects on FA and cellular features.
To explore the biological pathways that regulate the various FA and cellular features, either separately or together, the “hit siRNAs” were clustered according to their phenotypes. To further analyze the effects of groups of genes on the various stages of cell adhesion (namely, FA establishment, FA maturation, and cell spreading), the hits were clustered according to their effects on each separate group of features (FA morphology, FA distribution, or cell shape). To search for enrichment of biological features in the clusters, we used Genomica software (Segal et al., 2004; http://genomica.weizmann.ac.il/), searching in the Human BioCarta and Human Gene ontology (biological process and molecular function) databases.
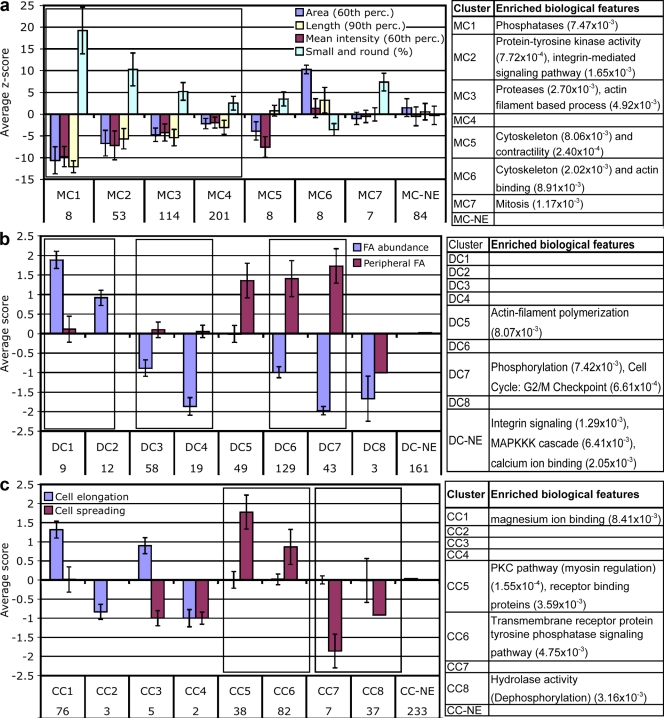
Three groups, each consisting of 8–9 clusters, were identified, some of which were enriched with a particular set of biological features. As shown in Fig. 4, each cluster was characterized by a particular “phenotypic signature,” representing the overall effect of the particular siRNAs. It is noteworthy that the graphs presented in Fig. 4 correspond to the mean strength of the effect of the siRNAs (according to the automatic z scores or manual scores). Thus, some of the clusters (e.g., MC1, -2, -3, and -4) could be regarded as having similar effects, though at varying degrees. These related clusters are grouped together in Fig. 4.
Figure 4.
Phenotypic signatures defined by clustering of the siRNA effects, and the biological features enriched in each cluster. Graphic representation of the mean scores of recorded features, showing the “phenotypic signature” of each cluster. The number of siRNAs in each cluster is indicated below the cluster name. Clusters with similar effects but with varying intensities are framed together. Tables on the right indicate biological features that are enriched in each cluster, with P values < 0.01. NE, no effect. Error bars represent standard deviation. (a) Clusters of FA morphology features. (b) Clusters of FA distribution features. (c) Clusters of cell shape features.
Clusters of FA morphological features (MC).
The most common effects of the siRNAs on FA morphology were characterized by inhibition of FA area, mean intensity, and length, and an increase in small and round FAs in varying degrees. Those effects did not change the correlation of the features, also shown in control cells. (Fig. 4 a; compare clusters MC1, -2, -3, and 4). However, the siRNAs associated with clusters MC5, -6, and -7 altered this tight correlation (Fig. 4 a). Clusters MC1 and MC2, which led to higher z scores, were mainly enriched with siRNAs targeting signaling molecules, whereas MC3, characterized by lower z scores, was enriched with siRNAs targeting proteases and actin filament-based processes. Cluster MC4 included siRNAs that induced slight changes in FA morphology; it also included a large number of siRNAs, the majority of which led to major changes in features represented in the other cluster sets. Clusters MC5 and MC6 were found to be enriched with cytoskeletal regulators.
Our analysis suggests that clusters MC1–4 primarily regulate the extent to which a preset assembly process is activated, whereas clusters MC5 and MC6 affect the individual structural features of FAs in varying ways. Cluster MC7, which induces an increase in small and round FAs (without affecting other morphological features) is enriched in mitosis-regulating genes. The “no-effect” (MC-NE) cluster contained all the siRNAs that had no significant effect on FA morphology, yet affected other features.
Clusters of FA distribution features (DC).
The FA distribution parameters (“DC clusters”) refer to the abundance and subcellular localization of the adhesion sites. Only 21 siRNAs induced an increase in FA abundance; DC1 and DC2 displayed different levels of essentially the same phenotype (Fig. 4 b). A striking result of this analysis may be seen in the high negative correlation between FA abundance and enrichment in peripheral FAs. Out of the 252 siRNAs from the low abundance FA clusters (DC3, -4, -6, -7, and -8), 172 led to enrichment in peripheral FAs, 77 did not change the FA distribution, and only 3 led to more centrally located FAs. These results are in accordance with previous results, showing that FAs are preferentially formed in the cell periphery (Izzard and Lochner, 1980; Zamir et al., 2000).
Those siRNAs inducing the formation of peripheral FAs (the DC5 cluster) were enriched in targeted genes involved in actin polymerization, whereas those leading to very low amounts of peripheral FAs (DC7) targeted genes involved in phosphorylation and in the G2/M cell cycle checkpoint.
Cell shape clusters (CC).
The cell shape parameters (“CC clusters”) included cell spreading (based on projected cell area) and cell elongation. The siRNAs that increased cell elongation (CC1) were biologically enriched in magnesium ion–binding targeted genes (Fig. 4 c). The three siRNAs in CC2 leading to low cell elongation were kinases. CC5, characterized by extensive spreading of largely nonelongated cells, contained siRNAs enriched in the myosin regulatory pathway, as well as receptor-binding signaling proteins. CC6, characterized by moderate cell spreading, was enriched with transmembrane receptor protein tyrosine phosphatases. The siRNAs leading to poor cell spreading (CC7) were rather diversified, whereas the siRNAs inducing mild cell contraction (CC8) were enriched with targeted genes involved in dephosphorylation.
A more detailed list of biological enrichments is shown in Tables S3, S5, and S7 for MC, DC, and CC clusters, respectively. The siRNAs belonging to each of the clusters are listed in Tables S4, S6, and S8, respectively.
Cross-correlation between different FA and cell shape features after siRNA perturbation
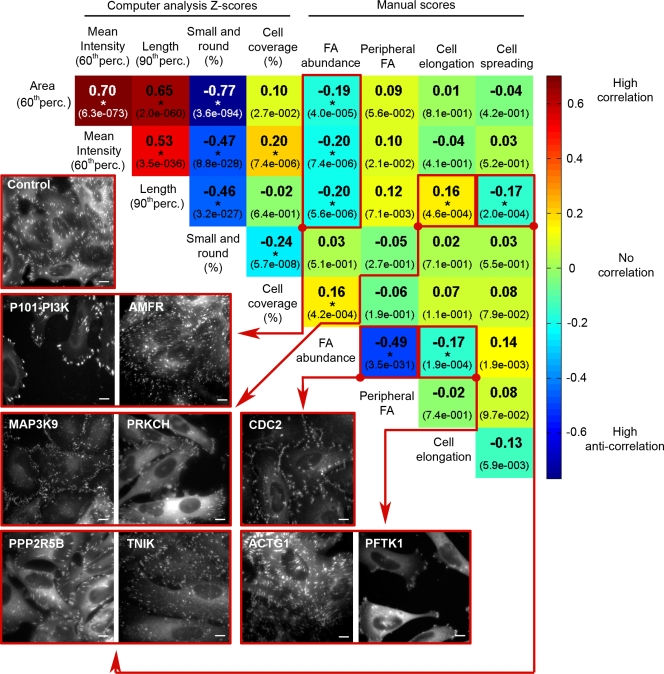
To determine which of the FA and cellular features were coregulated by the genes knocked down in this screen, correlations between pairs of measured features were calculated for all siRNA hits. Fig. 5 shows all the correlations found (significant correlations are marked with an asterisk), and representative images of the affected cells. As in the clustering analysis (Fig. 4 a), this analysis confirmed that FA morphological features were highly correlated in the screen hits, similar to their correlation in control cells. This finding indicates that only a small fraction of the siRNA hits (∼22%) interfered with these intrinsic correlations. These findings also demonstrated that cell coverage was positively correlated with FA mean intensity and abundance, and negatively correlated with the “small and round FAs” feature. Moreover, the higher the abundance of FAs, the greater the overall surface coverage by the cells.
Figure 5.
Cross-correlations between all pairs of FA features, cell shape features, and cell coverage, in the significant “hits” of the siRNA screen. Spearman correlation values (see Materials and methods) between all pairs of measured features were calculated for all the screen hits (483 siRNAs). Correlation values are color-coded (see scale bar on the right), and the corresponding correlation (R) value is designated in each square. P-values of the correlations are indicated by the numbers in parentheses. The images shown exemplify significant correlations (P < 0.0005; marked by an asterisk), and refer to the square or squares enclosed by the red line, and connected with an arrow from the red dot. For example, the negative correlation between FA abundance and FA area, mean intensity, and length is indicated by the low abundance of big, bright, and long FAs in P101-PI3K, and high abundance of small, faint and short FAs in AMFR. An image of control (RF) cells is shown for comparison. The siRNAs are designated by the siRNA-targeted Entrez gene symbol. Perc., percentile. Bars, 10 µm.
However, FA abundance was negatively correlated with FA area, mean intensity, and length. Indeed, in many cases, siRNA perturbation led to a decrease in the amount of FAs, and to an increase in FA intensity and length (e.g., TLN1, P101-PI3K). The opposite phenotype; namely, formation of many small, faint, and short FAs, was induced by another set of siRNAs (e.g., AMFR, CDC25C). These results suggest that the establishment, growth, and maturation of FAs are differentially regulated by specific sets of genes, enabling cells to respond to matrix interactions either by enlarging existing adhesions or by creating new ones.
Particularly intriguing were those siRNAs that break the “common correlations,” such as ACTG1 and FMN1, which induce highly abundant, yet normal or even large FAs; or APC and DUSP5, which promote the development of small, faint, and sparse FAs. Another interesting effect of siRNA is the perturbation of the common correlation between cell shape and FA morphology features. Thus, elongated cells tend to form longer FAs, whereas highly spread-out cells are characterized by shorter adhesions. This apparent linkage is specifically perturbed by such siRNAs as TLN1 and CSK (see Fig. S3 for a summary of siRNAs that uncoupled the feature correlations).
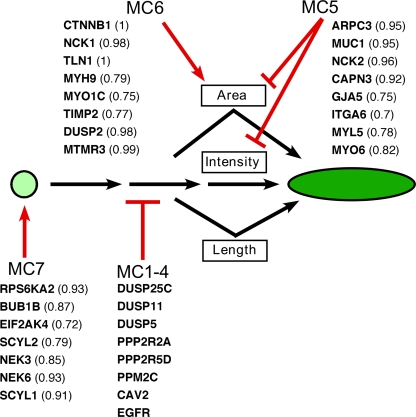
Our findings of correlations between FA morphological features suggest that there is a common molecular pathway regulating FA size, geometry, and paxillin content as small focal complexes mature into large FAs (Fig. 6). The siRNAs of clusters MC1–4 perturb this common pathway. Nevertheless, the coregulation of these features is not absolute, and may be modulated by specific siRNAs, which apparently “uncouple” these features. For example, siRNAs of the MC7 cluster enhanced the formation of dot-like adhesions without blocking the formation of “mature FAs”; MC6 enhanced FA area without affecting its elongation and intensity; and MC5 interfered with paxillin recruitment and FA area without affecting FA elongation. These findings point to a possible hierarchy in the distinct stages of FA development, with sequential regulation of individual features by distinct gene families (Fig. 6).
Figure 6.
Proposed hierarchy of FA morphology regulation. A proposed model for the hierarchy of genes regulating FA morphology. FA growth was found to be regulated by a common molecular pathway, leading to a coordinated increase in the FA's area, length, and paxillin concentration (indicated by the shape and green color intensity). This pathway was inhibited by siRNAs from clusters MC1–4 (the list of siRNAs in this figure is from MC1, which displayed the most striking effect). The concerted regulation of this pathway could be uncoupled by siRNA perturbations, which suggests additional, independent control of individual FA morphology features, as depicted. MC5 siRNAs inhibited the increase in the area and intensity of FA, without disturbing the increase in FA length. MC6 siRNAs affected the pathway, causing an increase in FA area, and MC7 siRNAs increased the percentage of small and round FAs, without affecting paxillin intensity. The siRNAs in the clusters that uncoupled the correlated features are shown together with their “uncoupling factor” in parentheses.
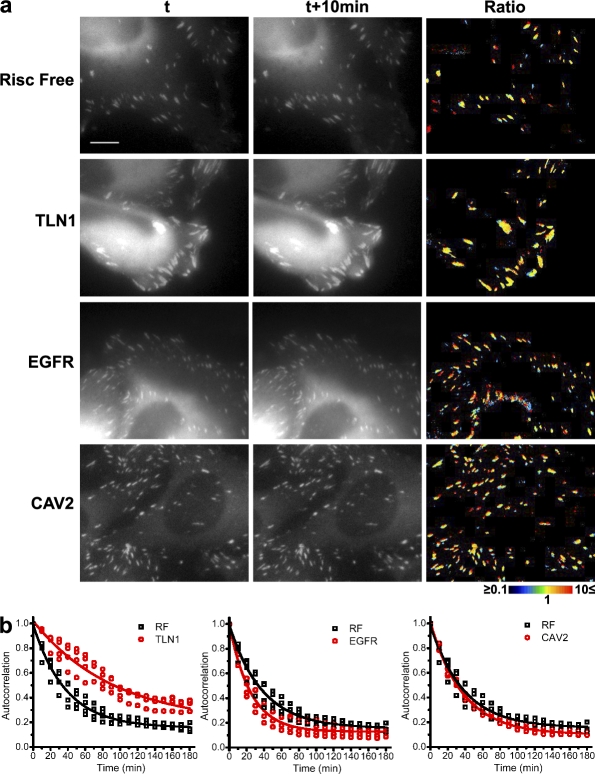
To assess this interpretation, we selected three siRNA hits with robust effects and, using time-lapse video microscopy, checked their effects on FA dynamics. TLN1 siRNA from cluster MC6, which reduces the number of FAs and increases their size and intensity compared with the RNA-induced silencing complex (Risc)-free control (RF), was found to display a dramatic reduction in FA initiation and turnover (Fig. 7 and Videos 1 and 2 for RF; and Videos 3 and 4 for TLN1). However, siRNAs of EGF receptor (EGFR) and CAV2, both of which perturb the “main pathway” of FA maturation, exert their effect via two distinct mechanisms. EGFR siRNA induces fast turnover of FAs, which suggests that the EGFR is involved in FA stabilization (Fig. 7 and Videos 5 and 6). CAV2 siRNA, however, induces FAs that are as stable as those of control cells, but their growth is prematurely arrested, which suggests the involvement of CAV2 in FA maturation (Fig. 7 and Videos 7 and 8). The knockdown efficiency of TLN1, EGFR, and CAV2 siRNAs is shown in Fig. S4 b.
Figure 7.
FA adhesion dynamics of TLN1, EGFR, and CAV2. Time-lapse videos (Videos 1–8) of HeLa cells transfected with TLN1, EGFR, CAV2, or Risc-free (RF) siRNA were used to create temporal ratio images (a) and for autocorrelation analysis (b). For each siRNA, the ratio between two time points, 10 min apart, is presented in color code: new pixels are blue, pixels that disappeared are red, and unchanged pixels are represented by variable colors, depending on the specific local intensity ratio. Yellow indicates identical intensities in the two time points. Note the high predominance of yellow pixels in TLN1-transfected cells, and note the high proportion of red and blue pixels in EGFR-transfected cells. For each siRNA, four different videos were used to perform autocorrelation analysis. Autocorrelation is calculated by comparing the adhesions at each time point with those at time zero. All points are plotted, as well as the single exponential decay fitting curve. The extracted decay time is: RF = 37.81 ± 2.07 min, TLN1 = 94.25 ± 12.71 min, EGFR = 24.45 ± 1.32 min, and CAV2 = 37.50 ± 0.89 min. The differences between RF, TLN1, and EGFR are highly significant (P < 0.0001). The knockdown of the three siRNAs is shown in Fig. S4 b. Bar, 10 µm.
Validation of the phenotypes induced by siRNA hits
To validate the effects of the siRNAs, a group of 86 siRNAs, displaying the most prominent effects, were selected. These siRNAs were chosen based on the phenotypic clustering data, with priority given to siRNAs from different clusters, especially those displaying high z scores, phenotypes that uncouple the normal feature correlations, or those that induce rare phenotypes (Table S9). The validation itself involved screening for the cellular effects induced by the corresponding ON-TARGETplus (OTP) pools. OTPs have been shown to yield a reduced frequency of off-target effects (Jackson et al., 2006), but were shown to produce weaker phenotypes (Simpson et al., 2008); therefore, we used them at a concentration of 100 nM. We scored the effects as in the primary screen, and also compared the phenotypes visually. The siRNAs in which most of the features were similar to the SMARTpool phenotypes were designated as validated.
We then assessed the knockdown efficiency of eight siRNAs that were chosen for validation. We did not see a significant difference in the knockdown efficiency between the SMARTpool reagents and the OTP reagents, even in the nonvalidated siRNA phenotype (PHB; Fig. S4 b), which indicates that knockdown efficiency is not the only reason for the failure in validation. 44 siRNAs were validated (sample images of selected validated hits are shown in Fig. S4 a). Table I summarizes the validated siRNAs, together with their cluster assignment. The genes belonging to the adhesome, and whether they are intrinsic (FA component [FAC]) or associated (FA-associated component [FACA]) to FAs, are indicated. We also summarized their relevance to adhesion, cytoskeleton, FA structure, and cell migration, according to gene ontology annotations (supported by experimental evidence). 22 of our validated hits were not previously associated with adhesion and migration.
Table I.
List of validated hits
| Gene ID | siRNA | Alias | Clusters | Adhesome | GO | ||
| FA morphology | FA distribution | Cell shape | |||||
| 5217 | PFN2 | Profilin 2 | MC-NE | DC7 | CC7 | NR | NR |
| 9829 | DNAJC6 | DJC6 | MC4 | DC-NE | CC4 | NR | NR |
| 3706 | ITPKA | IP3KA | MC-NE | DC7 | CC1 | NR | NR |
| 4690 | NCK1 | NCK-α | MC6 | DC5 | CC-NE | NR | Cytoskeleton |
| 55742 | PARVA | MXRA2 | MC4 | DC7 | CC1 | FAC, cytoskeletal | FAC, adhesion |
| 5578 | PRKCA | PKC-α | MC3 | DC6 | CC6 | FAC, S/T kinase | NR |
| 545 | ATR | FRP1 | MC-NE | DC7 | CC1 | NR | NR |
| 29904 | EEF2K | eEF-2K | MC-NE | DC7 | CC2 | NR | NR |
| 2870 | GRK6 | GPRK6 | MC-NE | DC7 | CC6 | NR | NR |
| 3685 | ITGAV | CD51 | MC2 | DC-NE | CC1 | FAC, adhesion | FAC, adhesion |
| 4646 | MYO6 | DFNA22 | MC5 | DC6 | CC1 | NR | Cytoskeleton |
| 5613 | PRKX | PKX1 | MC3 | DC6 | CC5 | NR | NR |
| 157 | ADRBK2 | BARK2 | MC-NE | DC7 | CC-NE | NR | NR |
| 369 | ARAF | A-RAF | MC3 | DC6 | CC5 | NR | NR |
| 998 | CDC42 | CDC42Hs | MC-NE | DC-NE | CC3 | NR | Cytoskeleton |
| 1847 | DUSP5 | DUSP | MC1 | DC6 | CC-NE | NR | NR |
| 9294 | EDG5 | S1PR2 | MC-NE | DC7 | CC1 | NR | NR |
| 10082 | GPC6 | MGC126288 | MC2 | DC5 | CC7 | NR | NR |
| 9270 | ITGB1BP1 | ICAP1 | MC-NE | DC7 | CC3 | NR | FAC, adhesion, migration |
| 79834 | KIAA2002 | SGK269 | MC3 | DC1 | CC-NE | NR | NR |
| 78986 | DUSP26 | MKP8 | MC3 | DC7 | CC3 | NR | NR |
| 4641 | MYO1C | NMI | MC6 | DC4 | CC8 | NR | Cytoskeleton |
| 5289 | PIK3C3 | Vps34 | MC4 | DC6 | CC5 | NR | NR |
| 5310 | PKD1 | PBP | MC-NE | DC7 | CC1 | FAC, channel | FAC, adhesion |
| 5881 | RAC3 | RAC3 | MC3 | DC5 | CC1 | Cytoskeleton | |
| 6093 | ROCK1 | P160ROCK | MC3 | DC6 | CC5 | FACA, S/T kinase | Cytoskeleton, adhesion, migration |
| 6198 | RPS6KB1 | p70(S6K)-α | MC4 | DC7 | CC-NE | NR | NR |
| 7077 | TIMP2 | CSC-21K | MC6 | DC6 | CC-NE | NR | NR |
| 8491 | MAP4K3 | GLK | MC-NE | DC6 | CC8 | NR | NR |
| 81629 | TSSK3 | STK22C | MC4 | DC6 | CC1 | NR | NR |
| 8440 | NCK2 | NCKβ | MC5 | DC5 | CC1 | FAC, adapter | Cytoskeleton |
| 1956 | EGFR | ERBB1 | MC1 | DC2 | CC6 | NR | Adhesion, migration |
| 4637 | MYL6 | ESMLC | MC3 | DC8 | CC8 | NR | Cytoskeleton |
| 657 | BMPR1A | ALK3 | MC4 | DC7 | CC8 | NR | NR |
| 1844 | DUSP2 | PAC1 | MC6 | DC-NE | CC-NE | NR | NR |
| 2534 | FYN | SYN | MC3 | DC1 | CC-NE | FACA, Tyr kinase | NR |
| 5526 | PPP2R5B | PR61B | MC-NE | DC3 | CC7 | NR | NR |
| 26191 | PTPN22 | LYP | MC2 | DC7 | CC-NE | NR | NR |
| 858 | CAV2 | CAV | MC1 | DC-NE | CC-NE | NR | NR |
| 3480 | IGF1R | CD221 | MC-NE | DC7 | CC8 | NR | Migration |
| 6251 | RSU1 | RSP-1 | MC4 | DC7 | CC1 | NR | Adhesion |
| 7094 | TLN1 | TLN | MC6 | DC7 | CC7 | FAC, cytoskeletal | FAC, adhesion, migration |
| 9641 | IKBKE | IKKE | MC2 | DC-NE | CC5 | NR | NR |
| 5218 | PFTK1 | PFTAIRE1 | MC4 | DC4 | CC1 | NR | NR |
The list includes the validated siRNA hits and their “cluster assignment,” based on the screen results. The standard gene symbol is indicated alongside one commonly used alias (from Entrez gene database, National Center for Biotechnology Information). The genes belonging to the adhesome are indicated, together with the family type. The gene ontology (GO) column summarizes the relevance to adhesion, cytoskeleton, FA structure, and cell migration, according to GO annotations (supported by experimental evidence). NR, not relevant, meaning that the particular gene is not listed in the adhesome or does not have GO annotations related to FA, cytoskeleton, adhesion, or migration.
Discussion
In the high-resolution siRNA perturbation screen described herein, we probed the involvement of specific gene families in multiple aspects of integrin adhesion, as well as cellular morphogenesis. We primarily searched for genes that regulated specific morphological features of FAs, yet did not block the formation of matrix adhesions altogether (of the total 1,180 siRNAs screened, only 39, mostly kinases, dramatically reduced the number of adherent cells).
The choice of specific siRNA libraries for modulation of FA features was primarily motivated by recent information concerning the molecular constituents of integrin adhesions. In a recent article, we demonstrated that the “integrin adhesome,” which consists of the components and regulators of integrin adhesions, is highly enriched with phosphorylation and dephosphorylation events (Zaidel-Bar et al., 2007a). The adhesome itself is enriched with kinases and phosphatases, and many of its signaling and scaffolding components have been shown to be phosphoproteins (Zaidel-Bar et al., 2007a). Our screen contained 109 molecules from the adhesome, including all of the integrins. Of those, 54 were hits. In the validated group of 44 siRNAs, 8 are adhesome components: ITGAV, NCK2, FYN, PARVA, PKD1, PRKCA, ROCK1, and TLN1. Notably, siRNAs targeting ITGAV, PKD1, PARVA, and NCK2 led to changes in cell shape (elongated cells, CC1); the TLN1, PKD1, PARVA, and NCK2 phenotypes result in low abundant and peripheral FAs (DC6-7). These adhesome-intrinsic components led to profound effects in FA distribution and cell shape, but did not affect the FA morphology. On the contrary, the three protein kinases—FYN, PRKCA and ROCK1—affected the main pathway of FA development (cluster MC3). Two adhesome components led to low cell coverage: SYK and TRPM7. Both genes were previously shown to be involved in cell survival (Inatome et al., 2001; Wykes et al., 2007).
Surprisingly, the hits discovered in this screen were not particularly enriched in adhesome components, which suggests that the adhesome is regulated not only by its intrinsic, dedicated components, but also by additional, “upstream” phosphorylation-regulated processes. The high percentage of hits that resulted from our screen indicates that the phosphorylation state of a large variety of components affects the adhesome, resulting in morphologically diverse adhesion phenotypes.
Our two major objectives in this screen were: (i) to delineate the interrelationships between the various quantified features of FA, and (ii) to assign specific genes to the regulation of each feature or group of features. Our findings point to a possible hierarchy in the distinct stages of FA development. Accordingly, we propose a hierarchical model for FA development in which there is a common pathway of FA size, geometry, and paxillin content, and in which there are some gene families that uncouple these correlated features (Fig. 6). Notably, the siRNAs that uncoupled the correlations were those targeting cytoskeletal genes, whereas those siRNAs affecting the features in a concerted manner primarily target signaling proteins (Figs. 4 a and 6).
Another interesting observation points to a high correlation between small numbers of FAs and their tendency to localize at the cell periphery (siRNA clusters DC6 and -7). Given the fact that FA assembly is usually initiated at the cell periphery, and that FAs “migrate” centripetally (Izzard and Lochner, 1980; Zamir et al., 2000), we propose that genes included in these clusters might be involved in the regulation of FA lifespan, rather than in their initiation or migration. These relationships could be uncoupled by siRNA treatment (e.g., cluster DC8), which preferentially reduced peripheral adhesions, suggesting that the suppression of the corresponding genes leads to arrest of FA formation or stability.
The findings reported herein may be considered at two distinct levels. At the “systems” level, it appears that despite their seemingly robust appearance, adhesion sites are highly regulated structures, in which the various structural features can be individually modulated by specific scaffolding and signaling components. Nevertheless, with the exception of the relatively few siRNAs that induced nearly complete loss of cells, the effects observed here were moderate, and did not block FA formation altogether. This finding is in line with the notion that the adhesome is a highly interconnected network (Zaidel-Bar et al., 2007a). At the same time, examination of the effects of individual proteins on the dynamics of FA organization and formation revealed major changes in FA morphology, initiation, growth, and stability. This is manifested in the three siRNAs tested for their effect on the dynamics of FAs (TLN1, EGFR, and CAV2) affecting FA initiation, stability, turnover, and growth.
One of the most demanding aspects of this screen was the validation step, intended to unequivocally link the cellular responses to the elimination of one particular mRNA. The common validation approach used in high-throughput screens involves redundancy experiments, in which at least two if not multiple distinct silencing reagents targeting the same gene cause the same phenotype (Echeverri et al., 2006). This approach is only moderately effective for assessing multiparametric cellular effects. Based on our experience, the inherent variability in knockdown efficiency, the small variations in local cell density, and the effects of “gene-specific factors” (e.g., protein half-life, development of compensatory mechanisms, and duplex stability) suggest that a process that could accurately validate a multiparametric response should be individually optimized for each particular gene. In view of the apparent conflict between the assay's content and throughput, it was not surprising that only half of the hits were validated. We therefore chose to divide the hits we identified into three groups. (i) Genes whose silencing resulted in a statistically significant effect on at least one parameter; this group is, indeed, very large, corresponding to ∼45% of all siRNAs tested. This apparently high rate is to be expected, given the enrichment in the tested libraries of molecules known to have an overall effect on cell adhesion and cytoskeletal organization. (ii) A selected group of 86 siRNAs whose effects were most pronounced, and included the capacity to “uncouple” features that would otherwise be highly correlated. And finally (iii), a group of 44 validated hits.
When comparing our results with previous data on the targeting of specific genes, we found various examples that are in accordance with previous knockouts or inhibition phenotypes. Talin1 (TLN1) knockdown, for example, which led to poor cell spreading and low numbers of FAs in our screen, was shown to be an essential element in FA structure (Albigès-Rizo et al., 1995; Priddle et al., 1998; Nayal et al., 2004); its knockout in undifferentiated embryonic stem cells inhibited cell spreading and FA formation (Nuckolls et al., 1992) in a similar manner to our own knockdown phenotype.
ROCK1 is known to be a downstream effector of Rho, involved in contractility and in the growth of FA (Amano et al., 2000). The siRNA knockdown of Rho kinase (ROCK1) in our screen led to highly spread cells and small FAs, which would be expected from the impairment of cell contractility, and similar to the effects of a ROCK1 inhibitor (Mammoto et al., 2004).
Fibroblast cell lines derived from Nck1−/− Nck2−/− embryos display defects in cell motility and in the organization of the lamellipodial actin network (Bladt et al., 2003). Knockdown of NCK1 in our screen led to larger and peripheral FAs without affecting FA intensity or length, and had no effect on cell shape. Knockdown of NCK2, however, led to the formation of elongated cells with low numbers of peripheral FAs. Both phenotypes could lead to aberrant cell motility, though by different mechanisms.
The libraries screened in this work were also assayed in a wound-healing screen performed in the Brugge Laboratory (Harvard Medical School, Boston, MA). We compared our validated hits with the hits in the migration screen performed on MCF10A cells with the same libraries (Simpson et al., 2008). Although the two screens are very different (different cell line, wound healing vs. steady-state adhesion, and moderate cell density vs. monolayers), we found 13 hits in common. From the validated siRNA hits in our screen, five led to impaired cell migration (TLN1, RSU1, IFG1R, IKBKE, and PFTK1). The impaired cell migration of cells treated with TLN1 siRNA is in accordance with the low FA turnover that we observed (Fig. 7), indicating that TLN1 is required for FA turnover, which is essential for effective cell migration. RSU1 siRNA-treated cells displayed small numbers of small, faint, and round FAs in the cell periphery, and the cells acquired an elongated shape. RSU1, which is selectively up-regulated in basal high-grade breast tumors (Simpson et al., 2008), was shown to be localized to FAs, together with PINCH1 and ILK; moreover, an siRNA for RSU1 was previously reported to impair cell adhesion (Dougherty et al., 2005). The impairment of migration by RSU1 siRNA seems to be caused by the effect of RSU1 on cell adhesion.
In the migration screen, six of our validated hits led to accelerated closure of the wound in MCF10A cells (PFN2, DNAJ6, ITPKA, NCK1, PARVA, and PRKCA). siRNA targeting of PFN2 in our screen led to rounded cells with low abundant FAs at the cell periphery. Indeed, in the MCF10A migration screen, the PFN2 siRNA-treated cells also showed minimal adhesion, with erratic migration and no polarity. These findings show that even in very different systems, knockdown of key elements in the cell adhesion process results in profound effects on cell adhesion and migration.
EGFR and NCK2 lead to a low Alamar blue phenotype (indicating cytotoxicity or a reduction in cell number) in the migration screen, whereas in our screen, they led to remarkable adhesion phenotypes. The sensitivity to knockdown of these two genes is different between the cell lines.
A comparison between the lists of low coverage (FA screen) and low Alamar (migration screen) show that siRNAs targeting PLK1 and AURKA both resulted in low cell numbers, which is in line with the notion that both are kinases involved in cell cycle control (Taylor and Peters, 2008). Talin, which, in our screen, affected cell shape, was also found to affect cell shape in an siRNA screen using Drosophila cell lines (Kiger et al., 2003), indicating the conserved function of this protein.
Given the prominent effects obtained in this screen, we have placed images showing representative examples of the effects of all siRNAs tested in a database hosted by the Cell Migration Consortium (http://www.cellmigration.org/resource/discovery/geiger/geiger2009_rnai.cgi).
Collectively, these results provide the first set of data concerning genes regulating FA formation and imply novel hierarchical relationships previously unexplored.
Materials and methods
Reporter cell line
HeLa cells were retrovirally infected with YFP-tagged human paxillin in a pBabe vector. Single-cell cloning was used to obtain a morphologically uniform population.
SiRNA libraries
Libraries were obtained from Thermo Fischer Scientific (http://www.dharmacon.com/HomePage.aspx) based on gene annotations from 2004. SMARTpools consisted of mixtures of four different siRNA sequences targeting the same gene. Three human siARRAY siRNA Libraries (Thermo Fisher Scientific) were screened: a kinase library targeting 576 kinases (protein kinases, selected based on (Manning et al. [2002], and lipid kinases), a phosphatase library targeting 192 phosphatases, and a custom library targeting 312 expanded family members of genes with known or predicted roles in cell adhesion and migration.
Screening protocol
HeLa cells (250 cells per well) were plated in 50 µl DME (Invitrogen) + 10% FCS (Biological Industries) in 384-well plates (F-bottomed, μClear; Greiner Bio-One, GmbH), and cultured for 24 h at 37°C with 5% CO2. The next day, 30 µl of the culture medium was removed from the cells, and cells were then transfected by direct addition of the 5-µl transfection mixture (1.25 µl siRNA [2 µM or 1 µM] in 2.5 µl DME with 0.25 µl Oligofectamine [Invitrogen] in 2.5 µl DME). Transfection was performed in duplicate. Cells were incubated for 24 h, and 40 µl of DME was then added so that the cells were grown for two more days in DME with 3% FCS. Control wells were left either untransfected or transfected with siCONTROL Risc-free siRNA (Thermo Fisher Scientific). Each plate contained two wells with siCONTROL TOX (Thermo Fisher Scientific) as an internal transfection control. The siCONTROL TOX is an RNA duplex that, when transfected into the cell, leads to cell death. Transfection efficiency was assessed by estimating the amounts of cells that remained viable in the well. Only plates with >90% transfection were taken for the screen.
After treatment, cells were fixed in 3% PFA for 20 min, and washed with PBS. Plates with fixed cells in PBS were screened.
Two screens were performed at final siRNA concentrations of 100 nM and 50 nM.
Microscope and controlling program
The automated microscopy systems (Liron et al., 2006; Paran et al., 2006) were based on IX71 and IX81 microscopes (Olympus). Automation was provided by ProScan (Prior Scientific), and included an xy stage, focus, shutters, and excitation and emission filter wheels. A fast laser AutoFocus attachment (Liron et al., 2006) was applied to focus the objective before image acquisition by means of a charge-coupled device camera (Quantix 57; Photometrics). The system was controlled by self-written Resolve6D software running in the RedHat Linux operating system. University of California at San Francisco Image Visualization Environment (http://www.msg.ucsf.edu/IVE/) and Priism libraries were used for image display and graphic user interfaces (GUIs). The acquired images in each well were tiled into montages in order to enable an overall visual impression of cell variability and for detection of consistent effects.
Image analysis
The general design of our computerized image analysis was described previously (Paran et al., 2007). In brief, images were first segmented into objects, using a binary threshold for the high-pass filtered images. For each object, morphological, fluorescence intensity, and estimated background parameters were calculated. In addition, the percentage of area covered by cells related to total image area (“cell coverage”) was calculated for each image, using the difference between the cell-free background fluorescence level, and cytoplasm diffuse fluorescence. Object-by-object, multiparameter data were saved for each image in separate files. User-controlled ranges for each parameter (minimum, maximum, and gates) defined the objects to be included, and excluded outliers.
Data analysis
Scoring of FA and cellular features.
The statistical distribution of the morphological FA parameters in the cells is not normal (see Paran et al., 2006); therefore, the selected scoring approach involved calculation of “percentile values” of these features. For FA area and intensity parameters, the 60th percentile (i.e., the FA area below which 60% of all FAs are found) and the 90th percentile for FA length, which detects the presence of the relatively low abundant, long FAs, were scored. To evaluate simultaneous alterations in FA axial ratio and total area distribution, a two-parameter quadrant was evaluated, using the 30th percentile value of each parameter in control wells as the gate values (percentage of small and round FAs). The score was defined as the fraction of objects in the low axial ratio–low area quadrant, out of the total number of objects in the tested sample. Wells in which the mean cell coverage was low were filtered from the analysis, and the siRNAs producing this effect were grouped into the “low cell coverage” cluster.
Each plate contained 10–16 control wells. For each feature, i (i = 1:5), the mean Fi, and standard deviation Si of the control wells were calculated (excluding outliers), and a z score was assigned to each gene, defined as where is the value of feature i for siRNA j. Hits in our screen were those genes in which the z score was either higher than 3.5 or lower than −3.5. This corresponds to a false discovery rate of 0.001 (i.e., 1/1,000 of the hits are, on average, false hits). Our reported hits constitute those genes that scored as hits according to these criteria in both 50 nM and 100 nM siRNA concentrations, or those genes that were toxic at 100 nM and scored as hits at 50 nM.
Besides the FA morphological features that were automatically calculated, images were visually inspected, and four additional features were scored: two features describing FA distribution (namely, their abundance and their subcellular localization) and two features describing cell shape (elongation and spreading). All visual scores ranged from −2 to 2, where a value of 0 corresponds to control wells. The result of this analysis was a phenotype vector of length 8 for each siRNA.
To assess the reproducibility of our measurements, we calculated the mean coefficient of variance (standard deviation divided by the mean of all the z scores) for each siRNA in the screen.
Feature correlations.
Spearman correlations between all pairs of features of the screen hits (z scores and manual scores) were used for defining the relationships between phenotypic features. The application of the Spearman correlation, which uses feature ranks rather than absolute values, is more suitable than the Pearson correlation in our case, as some of our features were scored manually, and others were z scores, with different inherent scales. P-values constitute the probability of obtaining these correlations by random chance.
Assignment of “phenotypic signatures” to specific siRNAs.
Hits were hierarchically clustered using the Cluster 3.0 program (Euclidean distance, mean linkage), generating specific clusters of siRNAs that induce similar effects on the selected features (FA morphology, FA distribution, and cell shape). Analysis of biological feature enrichment was performed using Genomica software developed in E. Segal's laboratory (Weizmann Institute of Science, Rehovot, Israel; Segal et al., 2004; http://genomica.weizmann.ac.il).
To quantify the extent to which the phenotypes of specific siRNA deviate from the mean observed FA morphology feature correlations, we sorted the Mahalanobis (Mahalanobis, 1936) distance between the signature of each hit and the median signature, and ranked the resulting distances. Then, we calculated the percentile of the ranks for each siRNA. siRNAs with high percentiles of these ranks are those genes that uncouple the feature correlations (percentiles for the uncoupling siRNAs are shown in Fig. 6).
Time-lapse video microscopy and ratio imaging
Time-lapse movies were recorded using the Real Time Delta Vision System (Applied Precision, LLC), which consists of an inverted microscope (IX71; Olympus) equipped with a CoolSnap HQ camera (Photometrics) and weather station temperature controller (Precision Control, LLC), operated by SoftWoRx and Resolve3D software (Applied Precision, LLC). Images were acquired with a Plan-Apochromat 60×/1.40 NA objective (Olympus). Cells were maintained at 37°C in DME without Phenol Red and riboflavin (Biological Industries) and supplemented with 10% FCS under a humidified atmosphere of 5% CO2.
Temporal ratio images were calculated and presented as described previously (Zamir et al., 2000). In this process, the intensity ratio of identical pixels in images acquired at different time points is calculated and represented by a color scale, such that “new pixels” are blue, pixels that disappeared are red, and unchanged pixels are represented by variable colors, depending on the specific local intensity ratio. Yellow indicates identical intensities in the two time points. Autocorrelation analysis was performed as described previously (Zaidel-Bar et al., 2007b), comparing the relative intensities of all the image pixels at two time points. Temporal ratio images and autocorrelation analysis were performed on videos that were high-pass filtered for optimal visualization of FA, after determining a threshold above which all pixels were defined as parts of adhesion structures. The student's t test was performed with GraphPad Software, and graphs and fits were created with OriginPro 7.0 (OriginLab Corporation).
Online supplemental material
Fig. S1 shows images from control and selected (adhesion-related siRNA) wells. Fig. S2 shows the reproducibility of measured features. Fig. S3 shows examples of siRNAs that produce changes in correlated features, and siRNAs that break the correlations; the images of the siRNA-treated cells may be seen at http://www.cellmigration.org/resource/discovery/geiger/geiger2009_rnai.cgi. Fig. S4 shows representative images of the validated siRNA hits and knockdown measurements. Videos 1, 3, 5, and 7 show FA dynamics of the HeLa cells transfected with Risc-free, TLN1, EGFR, and CAV2 siRNAs, respectively. Videos 2, 4, 6, and 8 show the temporal ratios of the Risc-free, TLN1, EGFR, and CAV2 siRNAs, respectively. Table S1 shows the biological features enriched in the low-coverage cluster. Table S2 shows the screen results for all of the siRNAs tested. Tables S3, S5, and S7 describe the biological features enriched in all clusters, and the siRNA list of each cluster (MC, DC, and CC clusters) is presented in Tables S4, S6, and S8, respectively). Table S9 lists the prominent hits selected for validation, identifying their respective clusters and the validation result. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200901105/DC1.
Acknowledgments
We wish to thank Yaniv Lubling and Eran Segal (Weizmann Institute of Science, Rehovot, Israel) for their help with the Genomica software, Irena Lavelin for providing the YFP-paxillin plasmid, Yael Paran (National Institutes of Health Chemical Genomics Center, Rockville, MD) for helpful discussions concerning image analysis, William Pearson (University of Virginia, Charlottesville, VA) for the generation of an interactive website hosted by the Cell Migration Consortium, Kaylene Simpson and Joan Brugge (Harvard Medical School, Boston, MA, USA) for illuminating discussions and joint selection of siRNAs for the MAR library, and Barbara Morgenstern for editorial assistance.
This study was supported by a grant from the National Institutes of Health Cell Migration Consortium (U54 GM64346). B. Geiger holds the Erwin Neter Professorial Chair in Cell and Tumor Biology.
Footnotes
Abbreviations used in this paper: EGFR, EGF receptor; FAs, focal adhesions; FAC, FA component; FACA, FA-associated component; MAR, migration- and adhesion-related; OTP, ON-TARGETplus; RF, Risc-free control; ROCK, Rho kinase.
References
- Albigès-Rizo C., Frachet P., Block M.R. 1995. Down regulation of talin alters cell adhesion and the processing of the alpha 5 beta 1 integrin.J. Cell Sci. 108:3317–3329 [DOI] [PubMed] [Google Scholar]
- Amano M., Fukata Y., Kaibuchi K. 2000. Regulation and functions of Rho-associated kinase.Exp. Cell Res. 261:44–51 [DOI] [PubMed] [Google Scholar]
- Berrier A.L., Yamada K.M. 2007. Cell-matrix adhesion.J. Cell. Physiol. 213:565–573 [DOI] [PubMed] [Google Scholar]
- Bladt F., Aippersbach E., Gelkop S., Strasser G.A., Nash P., Tafuri A., Gertler F.B., Pawson T. 2003. The murine Nck SH2/SH3 adaptors are important for the development of mesoderm-derived embryonic structures and for regulating the cellular actin network.Mol. Cell. Biol. 23:4586–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell I.D. 2008. Studies of focal adhesion assembly.Biochem. Soc. Trans. 36:263–266 [DOI] [PubMed] [Google Scholar]
- Dougherty G.W., Chopp T., Qi S.M., Cutler M.L. 2005. The Ras suppressor Rsu-1 binds to the LIM 5 domain of the adaptor protein PINCH1 and participates in adhesion-related functions.Exp. Cell Res. 306:168–179 [DOI] [PubMed] [Google Scholar]
- Echeverri C.J., Perrimon N. 2006. High-throughput RNAi screening in cultured cells: a user's guide.Nat. Rev. Genet. 7:373–384 [DOI] [PubMed] [Google Scholar]
- Echeverri C.J., Beachy P.A., Baum B., Boutros M., Buchholz F., Chanda S.K., Downward J., Ellenberg J., Fraser A.G., Hacohen N., et al. 2006. Minimizing the risk of reporting false positives in large-scale RNAi screens.Nat. Methods. 3:777–779 [DOI] [PubMed] [Google Scholar]
- Geiger B., Bershadsky A., Pankov R., Yamada K.M. 2001. Transmembrane crosstalk between the extracellular matrix—cytoskeleton crosstalk.Nat. Rev. Mol. Cell Biol. 2:793–805 [DOI] [PubMed] [Google Scholar]
- Hynes R.O. 1992. Integrins: versatility, modulation, and signaling in cell adhesion.Cell. 69:11–25 [DOI] [PubMed] [Google Scholar]
- Inatome R., Yanagi S., Takano T., Yamamura H. 2001. A critical role for Syk in endothelial cell proliferation and migration.Biochem. Biophys. Res. Commun. 286:195–199 [DOI] [PubMed] [Google Scholar]
- Izzard C.S., Lochner L.R. 1980. Formation of cell-to-substrate contacts during fibroblast motility: an interference-reflexion study.J. Cell Sci. 42:81–116 [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K., et al. 2006. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing.RNA. 12:1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I., Krylyshkina O., Small J.V. 2002. Regulation of substrate adhesion dynamics during cell motility.Int. J. Biochem. Cell Biol. 34:746–761 [DOI] [PubMed] [Google Scholar]
- Kiger A.A., Baum B., Jones S., Jones M.R., Coulson A., Echeverri C., Perrimon N. 2003. A functional genomic analysis of cell morphology using RNA interference.J. Biol. 2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liron Y., Paran Y., Zatorsky N.G., Geiger B., Kam Z. 2006. Laser autofocusing system for high-resolution cell biological imaging.J. Microsc. 221:145–151 [DOI] [PubMed] [Google Scholar]
- Mahalanobis P.C. 1936. On the generalised distance in statistics.Proceedings National Institute of Science of India. 2:49–55 [Google Scholar]
- Mammoto A., Huang S., Moore K., Oh P., Ingber D.E. 2004. Role of RhoA, mDia, and ROCK in cell shape-dependent control of the Skp2-p27kip1 pathway and the G1/S transition.J. Biol. Chem. 279:26323–26330 [DOI] [PubMed] [Google Scholar]
- Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. 2002. The protein kinase complement of the human genome.Science. 298:1912–1934 [DOI] [PubMed] [Google Scholar]
- Nayal A., Webb D.J., Horwitz A.F. 2004. Talin: an emerging focal point of adhesion dynamics.Curr. Opin. Cell Biol. 16:94–98 [DOI] [PubMed] [Google Scholar]
- Nuckolls G.H., Romer L.H., Burridge K. 1992. Microinjection of antibodies against talin inhibits the spreading and migration of fibroblasts.J. Cell Sci. 102:753–762 [DOI] [PubMed] [Google Scholar]
- Paran Y., Lavelin I., Naffar-Abu-Amara S., Winograd-Katz S., Liron Y., Geiger B., Kam Z. 2006. Development and application of automatic high-resolution light microscopy for cell-based screens.Methods Enzymol. 414:228–247 [DOI] [PubMed] [Google Scholar]
- Paran Y., Ilan M., Kashman Y., Goldstein S., Liron Y., Geiger B., Kam Z. 2007. High-throughput screening of cellular features using high-resolution light-microscopy; application for profiling drug effects on cell adhesion.J. Struct. Biol. 158:233–243 [DOI] [PubMed] [Google Scholar]
- Priddle H., Hemmings L., Monkley S., Woods A., Patel B., Sutton D., Dunn G.A., Zicha D., Critchley D.R. 1998. Disruption of the talin gene compromises focal adhesion assembly in undifferentiated but not differentiated embryonic stem cells.J. Cell Biol. 142:1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry S.K., Burridge K. 2000. Focal adhesions: a nexus for intracellular signaling and cytoskeletal dynamics.Exp. Cell Res. 261:25–36 [DOI] [PubMed] [Google Scholar]
- Segal E., Friedman N., Koller D., Regev A. 2004. A module map showing conditional activity of expression modules in cancer.Nat. Genet. 36:1090–1098 [DOI] [PubMed] [Google Scholar]
- Simpson K.J., Selfors L.M., Bui J., Reynolds A., Leake D., Khvorova A., Brugge J.S. 2008. Identification of genes that regulate epithelial cell migration using an siRNA screening approach.Nat. Cell Biol. 10:1027–1038 [DOI] [PubMed] [Google Scholar]
- Taylor S., Peters J.M. 2008. Polo and Aurora kinases: lessons derived from chemical biology.Curr. Opin. Cell Biol. 20:77–84 [DOI] [PubMed] [Google Scholar]
- Wykes R.C., Lee M., Duffy S.M., Yang W., Seward E.P., Bradding P. 2007. Functional transient receptor potential melastatin 7 channels are critical for human mast cell survival.J. Immunol. 179:4045–4052 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Itzkovitz S., Ma'ayan A., Iyengar R., Geiger B. 2007a. Functional atlas of the integrin adhesome.Nat. Cell Biol. 9:858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R., Milo R., Kam Z., Geiger B. 2007b. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions.J. Cell Sci. 120:137–148 [DOI] [PubMed] [Google Scholar]
- Zamir E., Katz M., Posen Y., Erez N., Yamada K.M., Katz B.Z., Lin S., Lin D.C., Bershadsky A., Kam Z., Geiger B. 2000. Dynamics and segregation of cell-matrix adhesions in cultured fibroblasts.Nat. Cell Biol. 2:191–196 [DOI] [PubMed] [Google Scholar]