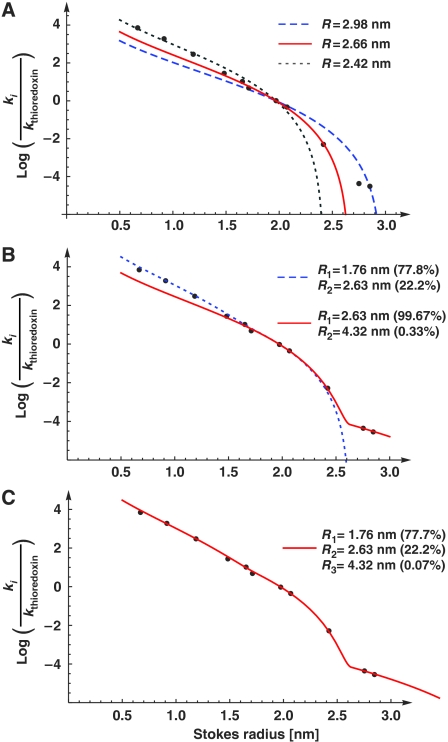
Figure 3.
Estimation of passive pore radii from passage rates. Graph shows a plot of Stokes radii (ri) versus the natural logarithm of ki/kthioredoxin, the ratios of nuclear entry rates. The following probes were included: a fluorescein maleimide-cysteine adduct (0.67 nm Stokes radius), a labelled 11 aa peptide (0.9 nm), insulin (1.19 nm), aprotinin (1.48 nm), profilin 1 (1.65 nm), a z-domain from Protein A (1.71 nm), thioredoxin (1.97 nm), lactalbumin (2.07 nm), GFP (2.42 nm), the phosphate-binding protein PBP (2.75 nm) and the maltose-binding protein MBP (2.85 nm). The data were parameter-fitted to different models for passive passage through NPCs (see equations (5) and (7)). A faithful model, consistent with all data points, could only be obtained with the assumption of heterogeneity in channel width. A smaller residuum indicates a better fit of the data (see below). For details see main text and Materials and methods. (A) ‘Uniformly sized channel model', as detailed in equation (5). The best fit was obtained for R=2.66 nm with a residuum of 3.58 (red curve). This fit, however, does not explain the very slow NPC passage of PBP and MBP. The panel also shows a simulated blue dashed curve with R=2.98 nm (residuum=5.87), which predicts best the flux ratio for MBP and thioredoxin. The gray dotted curve was obtained for R=2.42 (residuum=4.46); it predicts the flux ratios for small probes best. (B) ‘Two radii model' (with n=2 according to equation (7), Materials and methods). The radius combination of 1.76 and 2.63 nm gave a near-perfect fit for small and medium-sized probes (blue dotted line, residuum=2.75), whereas the combination 2.63 and 4.32 nm gave an excellent fit for medium sized and larger probes (red line, best global fit, residuum=1.51). (C) ‘Three radii model' (with n=3, equation (7)), which gave an excellent fit for small, medium sized and large probes (residuum=0.39).