Abstract
The success in profiling the phosphoproteome by mass spectrometry-based proteomics has been intimately related to the availability of methods that selectively enrich for phosphopeptides. To this end, we describe a protocol that combines two sequential enrichment steps. First, strong cation exchange (SCX) chromatography separates peptides by solution charge. Phosphate groups contribute to solution charge by adding a negative charge at pH 2.7. Therefore, at that pH, phosphopeptides are expected to elute earlier than their nonphosphorylated homologs. Second, immobilized metal affinity chromatography (IMAC) takes advantage of phosphate’s affinity for metal ions such as Fe3+ to uniformly enrich for phosphopeptides from the previously collected SCX fractions. We have successfully employed the SCX/IMAC enrichment strategy in the exploration of phosphoproteomes from several systems including mouse liver and Drosophila embryos characterizing over 5,500 and 13,000 phosphorylation events, respectively. The SCX/IMAC enrichment protocol requires 2 days, and the entire procedure from cells to a phosphorylation data set can be completed in less than 10 days.
INTRODUCTION
Reversible protein phosphorylation is a central regulatory mechanism of protein function in eukaryotes involved in countless cellular processes such as cell growth, cell differentiation, cell division and intercellular communication. Numerous efforts have been made aiming to understand how the phosphorylation state of a protein determines its conformation, activity and function, often on a single protein basis by mutation studies, kinase assays and 32P radiolabeling techniques1. While the information obtained from such highly focused studies is invaluable, a single phosphorylation event is often just a single part of an intricate signaling network linking many proteins and multiple phosphorylation sites. Furthermore, protein phosphorylation is believed to be a ubiquitous modification targeting most cellular proteins in one particular cellular state or another. Therefore, it is of great importance to study protein phosphorylation on a large scale to obtain a global picture of signaling events within the cell. Much of this can now be accomplished through mass spectrometry (MS)-based proteomics2.
The main difficulty in large-scale phosphorylation studies is the limitation in detecting phosphorylated species directly from complex sample mixtures due to the generally low expression levels of many regulatory phosphoproteins. Furthermore, phosphorylation often occurs at low stoichiometry, where the phosphorylated form of the proteinmay account for less than 1%of the total protein3. To solve these dynamic range issues, it is essential to perform an enrichment step that specifically selects for phosphorylated species. In this regard, the ability to survey the phosphoproteome has been intimately associated with the development of phosphopeptide-enrichment strategies.
Many enrichment strategies, normally performed at the peptide level, have proven successful. Peptide immunoprecipitation uses antibodies raised against a phosphorylation motif4 to isolate a particular subset of the phosphoproteome. IMAC enrichment utilizes metal cations such as Fe3+ (ref. 5) or Ga3+ (ref. 6) to coordinate phosphate groups by affinity, whereas titanium7,8 and zirconium9 oxides benefit from Lewis acid–base interactions. Chemical approaches to modify the phosphoester have also been proposed, using β-elimination and Michael addition10,11 or phosphoramidite chemistry12. Finally, ion (cation or anion) exchange chromatography separates peptides on the basis of solution charge. For SCX chromatography, phosphopeptides at acidic pH (~2.7) retain an additional negative charge and can be generally separated from nonphosphorylated peptides due to their reduced retention in the stationary phase13.
Additionally, one can combine separation techniques with phosphopeptide enrichment to reduce sample complexity and increase coverage. We have previously used SDS–polyacrylamide gel electrophoresis (SDS-PAGE) on protein whole cell extract and combined it with either IMAC or TiO2 at the level of peptides to identify around 2,000 and 3,000 phosphorylation sites from yeast Saccharomyces cerevisiae14 and Schizosaccharomyces pombe15, respectively. However, the method that has proven most comprehensive in mapping the phosphoproteome is SCX chromatography combined with either IMAC16–19 or TiO2 (refs. 20–22), and this method is the subject of our protocol. As phosphopeptide enrichment by TiO2 has been already described in detail in other protocols23,24, we will be focusing this one on SCX/IMAC. Nevertheless any of the two IMAC or TiO2 could be chosen in combination with SCX, and, in our hands, when working from the complexity obtained by collecting 10–15 fractions, both perform similarly. We believe superior results of SCX separation over gels are due to SCX chromatography grouping peptides on total charge and local charge distribution. Thus peptide mixtures to be enriched by IMAC or TiO2 are more similar in acidic properties, and competition occurs between species that are more alike, not favoring any phosphopeptide (e.g., multiple phosphorylated peptides) in particular.
The SCX/IMAC approach has allowed us to identify more than 5,500 phosphorylation sites in mouse liver18 and over 13,000 in fly embryos19 with relatively little effort and in less than 24 h of MS analysis time. The requirements we had in mind when developing this workflow were simplicity, robustness and a reduction in the MS analysis time to manageable levels, with a future view of automation and increased throughput. The protocol described here maintains the general outline used in previous studies, with a few added improvements toward selectivity and throughput of the IMAC enrichment.
Experimental design
Although the protocol can be adapted to special needs and available resources, in our experience, the following points reflect the setup providing best results:
Starting with enough material: at least 5 mg of protein, recommended 15 mg. Limited success has been observed with sample amounts <2 mg in terms of total number of phosphopeptide identifications.
Gradient for SCX chromatography should be expanded in the low charge region, where most phosphopeptides are expected to elute. This produces similar phosphopeptide complexity in all fractions. For tryptic digestions, this will be the+1/+2 region (see ANTICIPATED RESULTS for further details), for LysC digestions, steeper gradients should be used to expand +2 and +3 regions.
Use relatively fresh IMAC resin (to be determined batch to batch) and do not freeze and thaw the resin multiple times. The performance of these resins deteriorates with time.
Use an instrument capable of high mass accuracy measurements for precursor ions. This increases the number of phosphopeptide identifications by 40–50% (ref. 25) (see ANTICIPATED RESULTS for further details).
Run technical duplicates in the mass spectrometer to increase the number of phosphopeptide identifications by another 30–40%(refs. 19,25) (see ANTICIPATED RESULTS for further details). Additionally, experimental replicates are recommended for quantitative studies that somehow link phosphorylation dynamics to biological function.
MATERIALS
REAGENTS
Acetic acid (HAcO) (Ultrex II Ultrapure, J.T. Baker, cat. no. 6903)
Formic acid (FA) (EMD, cat. no. 11670)
Trifluoroacetic acid HPLC grade (TFA) (EMD, cat. no. TX1276)
Methanol HPLC grade (Burdick & Jackson, cat. no. AH230-4)
Acetonitrile HPLC grade (ACN) (Burdick & Jackson, cat. no. AH015-4)
Water HPLC grade (Burdick & Jackson, cat. no. AH365-4)
Sequencing grade modified trypsin (Promega, cat. no. V5111 or V5113)
Tris(hydroxymethyl)aminomethane (Tris) (Sigma, cat. no. T6066)
Sodium chloride (Sigma, cat. no. S6191)
Calcium chloride (Sigma, cat. no. C5670)
Urea (Sigma, cat. no. U6504)
Complete protease inhibitors cocktail tablets mini, EDTA-free (Roche, cat. no. 11 846 170 001)
Phenylmethylsulfonyl fluoride (PMSF) (Sigma, cat. no. P7626)
Sodium orthovanadate (Sigma, cat. no. S6508)
Sodium fluoride (Sigma, cat. no. S7920)
Sodium β-glycerophosphate (Sigma, cat. no. G6251)
Sodium pyrophosphate (Sigma, cat. no. S6422)
Bicinchoninic acid (BCA) protein assay kit (Pierce, cat. no. 23225)
Dithiothreitol (Biovectra, cat. no. 1369)
Iodoacetamide (Sigma, cat. no. I1149)
Glass beads 0.5-mm diameter (Biospec, cat. no. 11079105)
PhosSelect iron affinity gel (IMAC beads) (Sigma, cat. no. P9740) stored in aliquots at −20 °C
Potassium phosphate monobasic (Sigma, cat. no. P5379)
Potassium phosphate dibasic (Sigma, cat. no. P9666)
Phosphoric acid (J.T. Baker, cat. no. 7664-38-2)
Potassium chloride (Sigma, cat. no. P4504)
EQUIPMENT
HPLC system
Centrifuge
Vacuum manifold
Vortex
Lyophilizer
SpeedVac
LTQ Orbitrap mass spectrometer (Thermo Fisher) equipped with LC system and autosampler
REAGENT SETUP
Yeast cells
Yeast cultures should be grown under the confluence and media conditions desired for each particular experiment. Most wild-type S. cerevisiae strains yield around 100mg of protein per liter of culture at OD (600 nm)= 1.0.
Mammalian cells
Mammalian cells should be grown in the media required for each particular cell type. Most experiments are performed at 80% cell confluence, and protein amounts on the order of 1–5 mg are obtained from one 15-cm-diameter dish, depending on the cell type.
Lysis buffer for yeast
8 M urea, 75 mM NaCl, 50 mM Tris, pH 8.2, two tablets of protease inhibitors cocktail (complete mini, Roche) per 10 ml of lysis buffer, 50 mM NaF, 50 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1 mM PMSF.  It is recommended to prepare the buffer fresh before use. Concentrated stocks of phosphatase inhibitors (NaF, β-glycerophosphate, sodium orthovanadate and sodium pyrophosphate) can be stored at room temperature (18–24 °C). The storage temperature for 100 mM stocks of the protease inhibitor PMSF in DMSO or MeOH is −20 °C. PMSF should be added to the lysis buffer immediately before use, as it is unstable in aqueous solutions.
It is recommended to prepare the buffer fresh before use. Concentrated stocks of phosphatase inhibitors (NaF, β-glycerophosphate, sodium orthovanadate and sodium pyrophosphate) can be stored at room temperature (18–24 °C). The storage temperature for 100 mM stocks of the protease inhibitor PMSF in DMSO or MeOH is −20 °C. PMSF should be added to the lysis buffer immediately before use, as it is unstable in aqueous solutions.
Lysis buffer for mammalian cells or tissues
8 M urea, 75 mM NaCl, 50 mM Tris, pH 8.2, one tablet of protease inhibitors cocktail (complete mini, Roche) per 10 ml of lysis buffer, 1 mM NaF, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1 mM PMSF.  Use same recommendations as for the yeast lysis buffer.
Use same recommendations as for the yeast lysis buffer.
SCX buffers
(A) 7 mM KH2PO4, pH 2.65, 30% ACN (vol/vol); (B) 7 mM KH2PO4, 350 mM KCl, pH 2.65, 30% ACN (vol/vol); (C) 50 mM K2HPO4, 500 mM NaCl, pH 7.5.  As organic solvents affect the pH reading, the pH adjustments for buffers A and B should be performed before the addition of ACN.
As organic solvents affect the pH reading, the pH adjustments for buffers A and B should be performed before the addition of ACN.
IMAC buffers
Binding buffer: 40% ACN (vol/vol), 25 mM FA in H2O Elution buffer A: 50 mM K2HPO4, adjust to pH 10 with NH4OH Elution buffer B: 500 mM K2HPO4, pH 7.
SepPak solvents
Binding buffer: 0.1% TFA (vol/vol) in H2O Elution buffer: 50% ACN (vol/vol), 0.5% HAcO (vol/vol) in H2O.
StageTip solvents
Binding buffer: 1% FA (vol/vol) in H2O Elution buffer: 50% ACN (vol/vol), 0.5% HAcO (vol/vol) in H2O.
LC-MS/MS (Liquid chromatography coupled to tandem mass spectrometry) solvents
(A) 3% ACN (vol/vol), 0.125% FA (vol/vol) in H2O; (B) 0.125% FA (vol/vol) in ACN.
EQUIPMENT SETUP
HPLC system for SCX chromatography
Valco valve for manual injection set with a 500-µl loop, quaternary pump Agilent 1100 equipped with degasser, operating at 3 ml min−1, photodiode array (PDA) detector from ThermoFisher set up to read at fixed 220- and 280-nm wavelengths.
Semipreparative SCX column: polySULFOETHYL A, 9.4-mm inner diameter × 200 mm length, 5-µm particle size, 200 Å pore size (PolyLC).
LC-MS/MS equipment
Famos autosampler (Dionex) with 10-µl loop and2.4-µl injection needle, Agilent 1100 pumps operating between 80 and 300 µl min−1 and setup with a double-split system to provide an in-column flow rate of 0.5–1 µl min−1 on a microcapillary column 125 µm × 18 cm column with hand-pulled tip packed with C18 reverse-phase material (MagicC18AQ, MichromBioResources) and LTQOrbitrap mass spectrometer equipped with a nanospray source. The method used for acquisition consists of an 80-min method with 9-min loading time, 57-min gradient from 4% to 26% B (see REAGENT SETUP for solvent composition) and 60 min of MS data collection. For each scan cycle, one full MS scan acquired in the Orbitrap mass analyzer at 60,000 resolution, 1 × 106 automatic gain control (AGC) target and 1,000 ms maximum ion accumulation time is followed by 10 MS/MS scans on the ten most intense ions in the linear ion trap (LTQ) at 3,000 signal threshold, 5,000 AGC target and 120 ms maximum accumulation time, 2.0 Da isolation width and 30 ms activation at 29% normalized collision energy. Dynamic exclusion is enabled to exclude from fragmentation ions that had been already selected for MS/MS in the previous 35 s. Ions with a charge of +1 or unassigned are also excluded. All scans (MS and MS/MS) are collected in centroid mode. To ensure proper performance of the system, we use a complex mixture of yeast peptides corresponding to 1/10 of the yeast proteome and obtained by SDS-PAGE fractionation of cell protein extract and trypsin digestion of gel regions (see TROUBLESHOOTING).
PROCEDURE
-
1
Prepare the cell lysate using option A for yeast and option B for mammalian cultured cells.
-
Preparation of yeast lysate
 During lysis, cells and lysates should be kept at 4 °C at all times.
During lysis, cells and lysates should be kept at 4 °C at all times.Harvest the cells by centrifugation at 2,000g for 15 min at 4 °C. Wash the cell pellet with ice-cold water, divide into screwed-cap 2-ml tubes and centrifuge again at 10,000g for 5 min at 4 °C to remove the water. Ideally there should be 250 µl of dried cell pellet in each tube.
Add 700 µl of lysis buffer previously chilled at 4 °C and 1,400 µl of glass beads; 10–15% of the volume should remain as an air chamber for proper agitation.
Use the Mini Beadbeater (Biospec) for microcentrifuge tubes at maximum speed, three cycles of 90 s each, and waiting 2 min in between each cycle to avoid overheating of the lysates.
Poke a hole in the bottom of the tube, with a flame-heated 21-gauge needle and place the microcentrifuge tube on top of a 5-ml tube. Centrifuge to separate the lysate from the glass beads at 1,000g for 3 min at 4 °C. At this point, only the glass beads should remain in the microcentrifuge tube and all the lysate should have been transferred to the 5-ml tube; otherwise, repeat this step.
Homogenize the lysate by pipetting up and down, and transfer to 1.5-ml microcentrifuge tubes. Centrifuge at 15,000g for 10 min at 4 °C and transfer the supernatant into new tubes.
-
Measure protein concentration using Lowry, Bradford or BCA protein assay. For yeast lysates, the concentration should be approximately 10–20 mg ml−1.
 It is recommended to continue with Step 2 immediately to minimize protein degradation. If lysates need to be stored at this point, store them at −80 °C.
It is recommended to continue with Step 2 immediately to minimize protein degradation. If lysates need to be stored at this point, store them at −80 °C.
-
Preparation of mammalian cultured cells lysate
Wash cells from 15-cm dishes with 5-ml ice-cold PBS.
Aspirate PBS and add 1 ml of chilled lysis buffer covering entirely the cell monolayer and place dishes at 4 °C for 10 min.
Scrape cells and transfer lysates into 15-ml conical tubes.
Sonicate 3× 60 s at 4 °C with 2 min rest in between using output ~15%.
Centrifuge at 2,500g for 10 min at 4 °C to eliminate cell debris and transfer the supernatant into new tubes.
-
Measure protein concentration using Lowry, Bradford or BCA protein assay. For most mammalian cell lines, the concentration is expected to be approximately 1–3 mg ml−1.
 It is recommended to continue with Step 2 immediately to minimize protein degradation. If lysates need to be stored at this point, store them at −80 °C.
It is recommended to continue with Step 2 immediately to minimize protein degradation. If lysates need to be stored at this point, store them at −80 °C.
-
Protein reduction and alkylation
-
2
Add DTT from a 0.5 M stock to 5 mM final concentration and incubate for 25 min at 56 °C to reduce disulfide bonds.
 Avoid temperatures higher than 60 °C where urea-based carbamylation of lysines and protein N-termini can occur.
Avoid temperatures higher than 60 °C where urea-based carbamylation of lysines and protein N-termini can occur. -
3
Allow the protein mixture to cool to room temperature and add iodoacetamide to 14 mM final concentration. Incubate for 30 min at room temperature and in the dark to alkylate cysteines.
-
4
Quench unreacted iodoacetamide by adding 0.5 M DTT to additional 5 mM and incubating 15 min at room temperature in the dark. See Box 1.
CHECKPOINTS.
We recommend checking the sample at various stages of the protocol to ensure that every step is performed successfully. Here is a list of suggested checkpoints:
Run an SDS-PAGE gel after Step 4 to qualitatively determine protein integrity. Lack of proteins at the high molecular weight gel region implies protein degradation during lysis.
Collect a small aliquot from Step 16 and analyze by LC-MS/MS after removing ACN. Peptides should be observed throughout the gradient.
Analyze a few SCX fractions from several points in the gradient (e.g., fractions 3, 7 and 11) after desalting (Step 26). Peptides should again be observed throughout the gradient. Also, verify solution charge distribution of identified peptides and observe an increase in peptides with higher charges in the later fractions.
Trypsin digestion
-
5
Dilute the protein mixture 1:5 in 25 mM Tris-HCl, pH 8.2, to reduce the concentration of urea to 1.6 M.
-
6
Add CaCl2 from a 0.1 M stock to 1 mM.
-
7
Add trypsin at a minimum concentration of 4–5 ng µl−1 and 1/200-1/250 enzyme:substrate. Incubate at 37 °C overnight.
-
8
Allow the digest to cool to room temperature and stop the digestion by acidification with TFA to 0.4% (vol/vol). Verify that the pH is <2.0; otherwise add more acid.
-
9
Centrifuge at 2,500g for 10 min at room temperature and discard the pellet.
Peptide desalting
-
10
Peptides must be desalted before SCX analysis to remove salts and urea from the digestion buffer. We use reverse-phase tC18 SepPak solid-phase extraction cartridges from Waters. The size of the cartridge should be selected on the basis of the amount of starting protein, considering their capacities are about 5% (wt/wt) of the packing material’s weight. For example, for 20 mg of a protein digest, a SepPak with 500 mg of tC18 beads is recommended. In this protocol, we assume the use of 500 mg of SepPak (500-mg bulk material, 3 or 6 ml, 800-µl bed volume). Volumes should be adapted accordingly for different sizes. A vacuum manifold can be used to increase solvent flow rates through the cartridge, or alternatively, air pressure is recommended for high-capacity SepPaks, as it provides more uniform peptide loading and elution.
-
11
Wash and condition the cartridge using 9 ml of ACN followed by 3 ml of 50% ACN and 0.5% HAcO.
-
12
Equilibrate with 9 ml of 0.1% TFA
-
13
Load sample in 0.4% TFA.
-
14
Wash/desalt with 9 µl of 0.1% TFA.
-
15
Wash (to remove TFA) with 900 µl of 0.5% HAcO.
-
16
Elute with 5 ml of 50% ACN 0.5% HAcO and collect eluate in a 15-ml conical tube. See Box 1.
-
17
Freeze the eluate with liquid N2 and lyophilize. The result here should be a white (sometimes yellowish) fluffy powder.
 At this point, samples can be stored at −20 °C for several weeks.
At this point, samples can be stored at −20 °C for several weeks.
SCX chromatography
-
18
Equilibrate the SCX HPLC system by running a blank run. Set up the gradient described in Table 1 for peptide separation.
-
19
Resuspend lyophilized peptides in 400–500 µl of SCX buffer A and inject onto the HPLC system used for SCX chromatography. Collect twelve 4-min fractions from minute 1 to minute 49.
 For high-resolution chromatography, it is important to keep the amount of peptides loaded below the maximum column capacity. For the SCX column described here, the capacity is ~16 mg of tryptic peptides, but best results are accomplished with ~5 mg. Reproducible SCX chromatography is obtained with long and constant equilibration times.
For high-resolution chromatography, it is important to keep the amount of peptides loaded below the maximum column capacity. For the SCX column described here, the capacity is ~16 mg of tryptic peptides, but best results are accomplished with ~5 mg. Reproducible SCX chromatography is obtained with long and constant equilibration times. -
20
Freeze the fractions with liquid N2 and place them into the lyophilizer to eliminate the ACN. Fractions need not be completely dried for the next step; therefore, they can be used once the volume has been reduced by 30%.
TABLE 1.
Gradient used for separation of tryptic peptides by SCX chromatography with increased expansion of low (+1 and +2) solution charges.
| Time (min) |
Buffer A (%) |
Buffer B (%) |
Buffer C (%) |
H2O (%) |
|---|---|---|---|---|
| 0 | 100 | 0 | 0 | 0 |
| 2 | 100 | 0 | 0 | 0 |
| 35 | 75 | 25 | 0 | 0 |
| 36 | 0 | 100 | 0 | 0 |
| 41 | 0 | 100 | 0 | 0 |
| 42 | 0 | 0 | 0 | 100 |
| 48 | 0 | 0 | 0 | 100 |
| 49 | 0 | 0 | 100 | 0 |
| 60 | 0 | 0 | 100 | 0 |
| 61 | 0 | 0 | 0 | 100 |
| 67 | 0 | 0 | 0 | 100 |
| 68 | 100 | 0 | 0 | 0 |
| 120 | 100 | 0 | 0 | 0 |
Peptide desalting (II)
-
21
Peptides are again desalted to remove salts from the SCX buffers by reverse phase using solid-phase extraction cartridges as described above. We use 100 mg of tC18 SepPaks from Waters and a vacuum manifold. Wash and condition the cartridge with 3 ml of ACN and then with 3 ml of 50% ACN 0.5% HAcO.
-
22
Equilibrate with 3 ml of 0.1% TFA.
-
23
Load sample in 0.1% TFA.
-
24
Wash/desalt with 3 ml of 0.1% TFA.
-
25
Wash (to remove TFA) 250 µl of 0.5% HAcO.
-
26
Elute with 1 ml of 50% ACN 0.5% HAcO and collect the eluates in 2-ml Eppendorf tubes. See Box 1.
-
27
Freeze the eluates with liquid N2, and lyophilize or dry the peptides by vacuum centrifugation.
 At this point, sample can be stored at −20 °C for several weeks.
At this point, sample can be stored at −20 °C for several weeks.
IMAC phosphopeptide enrichment
-
28
Prepare 100 µl of IMAC beads by washing them with 1 ml of IMAC binding buffer, turning over the vial a few times to resuspend all beads and spinning to remove the liquid. Repeat three times and prepare 50% slurry in the same buffer.
-
29
Prepare twelve 250-µl Eppendorf tubes and place 10 µl of IMAC beads slurry into each. Cutting the end of the tip facilitates pipetting of beads.
-
30
Dissolve each peptide fraction obtained from Step 27 in 120 µl of IMAC-binding buffer and transfer to the IMAC beads obtained from Step 29.
-
31
Incubate peptides on beads for 60 min, with vigorous shaking, at room temperature.
-
32
During this time, prepare StageTips26 by packing two disks of Empore 3M C18 material onto 250-µl pipette tips.
-
33
Wash and equilibrate StageTips by passing through sequentially 20 µl of MeOH, 20 µl of 50% ACN 0.5% HAcO and twice through 20 µl of 1% FA. For convenience and increased throughput, one can use a centrifuge by holding the StageTips within a 2-ml Eppendorf tube with the top part of a 500-µl Eppendorf body as an adaptor, limiting spinning speed to 2,000g and time to the minimum to get the liquid passed through.
-
34
From this step, one can keep IMAC enrichment and phosphopeptide desalting separated (option A), or for convenience, experienced users might consider combining them into a single procedure (Option B). Both protocols are provided below.
-
IMAC enrichment followed by phosphopeptide desalting
Remove liquid from the tubes (Step 31) and transfer to a tube labeled ‘IMAC flowthrough’. These peptide mixtures can be analyzed in the mass spectrometer if one is interested in nonphosphorylated peptides.
Wash the resin with 120 µl of IMAC binding buffer. Repeat this step twice.
Elute phosphopeptides by incubating for 5 min with 40 µl of IMAC elution buffer A (50 mM K2HPO4/NH4OH, pH 10.0). Repeat this step twice. Combine eluates from the same sample into the same tube, containing 40 µl of 10% FA to neutralize pH.
Dry samples by vacuum centrifugation at room temperature.
Resuspend the samples from Step A(iv) in 20 µl of 1% FA and load them into the StageTips prepared on Steps 32 and 33.
Wash to desalt with 20 µl of 1% FA.
-
Combined IMAC enrichment and phosphopeptide desalting
After the 60-min incubation in Step 31, transfer IMAC beads to the top of the StageTips and spin down. The beads will get retained on the StageTip and the solution will pass through. As the buffer contains 40% ACN, nonphosphorylated peptides, which are not retained in the IMAC resin, will not be retained by the C18 material. These peptide mixtures can be collected and analyzed as well in the mass spectrometer.
Wash with 50 µl of IMAC binding buffer. Repeat this step once.
Wash with 40 µl of 1% FA. This step allows equilibrating the StageTip C18.
Wash with 70 µl of 500 mM K2HPO4, pH 7. Repeat this step twice. At this point, phosphopeptides are eluted from IMAC resin and retained on the C18.
Wash with 40 ml of 1% FA to remove phosphate salts.
-
-
35
Elute phosphopeptides from StageTips into vials for MS analysis with 40 µl of 50% ACN 0.5% HAcO.
-
36
Dry down the samples from Steps 34A(i) and 34B(i) (nonphosphorylated peptides) and Step 35 (phosphopeptides) by vacuum centrifugation.
 At this point, samples can be stored at −20 °C for several weeks.
At this point, samples can be stored at −20 °C for several weeks.
LC-MS/MS analysis
-
37
Resuspend samples in 10 µl of 5% ACN, 4% FA. For nonphosphopeptide samples, prepare a 1/100 dilution.
-
38
Inject 1 µl of each sample onto the LC-MS/MS system. See EQUIPMENT SETUP for details about configuration and settings. Analyze each sample in duplicate. Data analysis is discussed in ANTICIPATED RESULTS.
![]()
![]()
Cell lysis (yeast): cell harvesting and washing, 25 min; lysis, 30 min
Cell lysis (mammalian cells): cell harvesting and washing, 5 min; lysis, 35 min
Measuring protein concentration using BCA assay, 40 min
Protein reduction, alkylation and digestion: Steps 2–4, 70 min; Steps 5 and 6, 5 min; Step 7, overnight; Steps 8 and 9, 15 min
Peptide desalting: Steps 10–16, 30 min; Step 17 (lyophilize), 2–8 h (depending on volume)
SCX chromatography: Steps 18 and 19, 2 h; Step 20 (remove ACN by lyophilization), 3–4 h
Peptide desalting II: Steps 21–26, 30 min; Step 27 (lyophilize), 3 h
Combined IMAC enrichment and phosphopeptide desalting: Steps 28–30, 15 min; Steps 31–33, 1 h; Step 34A(i–iii), 1 h; Step 34A(iv), 2 h; Step 34A(v) and (vi), 15 min; Step 34B, 35 min; Step 35, 5 min; Step 36, 1 h
LC-MS/MS analysis: 80 min per run
![]()
Troubleshooting advice can be found in Table 2.
TABLE 2.
Troubleshooting table.
| Problem | Possible reason | Solution |
|---|---|---|
| No high molecular weight proteins after cell lysis |
Protein degradation | Make sure protease inhibitors were added before lysis. Avoid sample heating during lysis. Perform lysis steps up to protein digestion (Steps 1–7) the same day and as fast as possible |
| No peptides observed in checkpoint 2 (Box 1) |
Protein digestion failed | Check integrity/reactivity of trypsin solutions. pH during digestions should be 7.5–8.5 and urea concentration <2 M |
| Peptides elute too late when analyzed by LC-MS/MS |
Maximum capacity was reached on the SepPak cartridge used for peptide desalting |
Use higher-capacity SepPak cartridge or save the flow-through and washes and perform a second desalting step with them |
| Peptides elute too early in the SCX. Peptides with high solution charge are observed in the early fractions |
SCX column has not been equilibrated properly |
Equilibrate the SCX column for a longer time |
| SCX buffers A and B have pH > 3 | Make new buffers and ensure they have been properly prepared |
|
| SCX buffer A has higher concentration of cations |
||
| Many nonphosphorylated peptides (>50%) are detected after IMAC enrichment |
Too much IMAC resin was used for the amount of peptides |
Reduce the amount of resin |
| Incubation time of peptides with IMAC resin was too long |
Do not incubate for longer than 60 min | |
| IMAC resin was not properly washed before elution |
Add an extra wash step with IMAC-binding buffer (i.e., repeat Step 34A(ii) or 34B(ii)) |
|
| Very few total peptides were detected | Starting amounts of protein were not sufficient |
Although lower amounts might work, we recommend starting with at least 5 mg of protein |
| IMAC resin is too old | PhosSelect IMAC resin shelf life is less than 1 year. Store in small aliquots at −20 °C and do not freeze and thaw multiple times. When performance is decreased, replace it with a new batch (an SCX fraction prepared following this protocol can be used as a control) |
|
| There was a problem in the LC-MS/MS system |
Make sure LC-MS/MS system is working properly with a complex mixture standard (see EQUIPMENT SETUP) |
ANTICIPATED RESULTS
SCX/IMAC for the collection of large-scale phosphorylation data sets
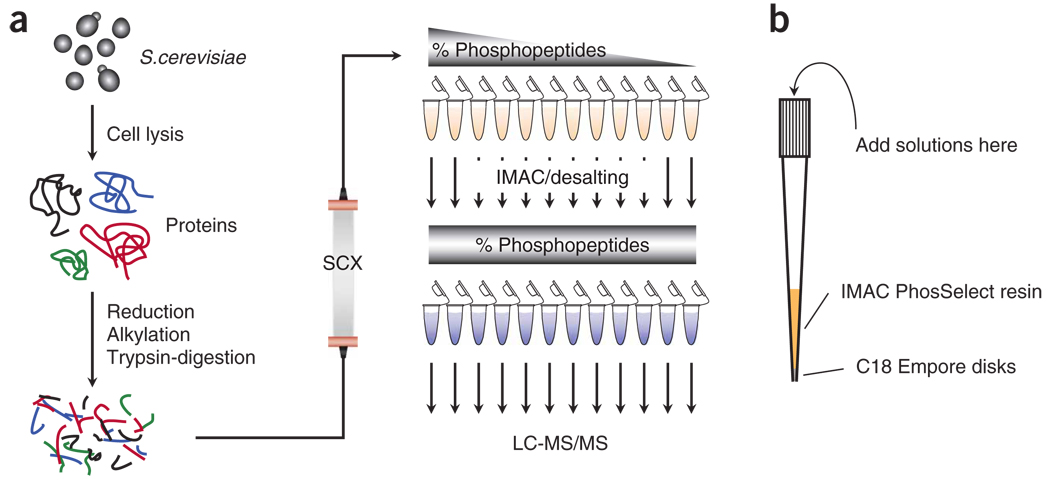
To exemplify the performance of the SCX/IMAC approach for large-scale phosphorylation studies, we applied this method to budding yeast S. cerevisiae. Figure 1a shows a schematic of the described workflow. Yeast were grown to mid-log phase, harvested and lysed. Fifteen milligrams of protein were reduced, alkylated and trypsin-digested. Desalted peptides were subjected to SCX chromatography and 12 fractions were collected, desalted, enriched by IMAC and desalted in one step (Fig. 1b), and analyzed by LC-MS/MS on an LTQ Orbitrap. All MS/MS spectra collected were searched with the Sequest algorithm27 against a composite yeast database with forward and reversed sequences28.
Figure 1. Diagrams illustrating the protocol.
(a) Scheme of the procedure for phosphopeptide enrichment and analysis. Protein extract is in-solution reduced, alkylated and digested with trypsin. Peptides are desalted and separated by SCX chromatography. Twelve fractions are collected, desalted, enriched by IMAC and analyzed by LC-MS/MS techniques. (b) Illustration of the StageTips used for combined IMAC enrichment and phosphopeptide desalting, where the IMAC resin, after incubation with peptide mixtures is finished, is packed on the top of C18 Empore disks. In the absence of ACN, phosphopeptides eluting from the IMAC resin are bound to the C18 disks, and salts can be removed before organic elution.
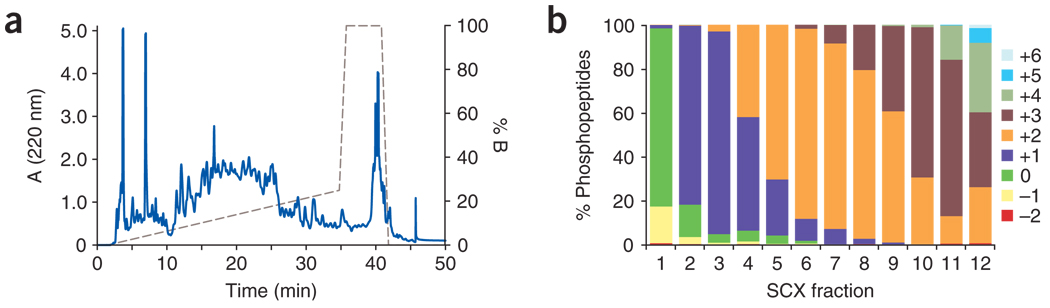
Separation of peptides at pH 2.65 by SCX chromatography is shown in Figure 2. The chromatogram of UV absorbance detection at 220 nm is displayed in Figure 2a, and the solution charge separation across the fractions of all the phosphopeptides detected after IMAC enrichment is shown in Figure 2b. The aforementioned buffer compositions and the gradient program we designed allow us to obtain increased separation between peptides with charges +1 and +2.
Figure 2. Performance of semipreparative SCX chromatography.
(a) Chromatogram of the SCX separation of peptides. (b) Solution charge separation of nonredundant phosphopeptides identified from each fraction.
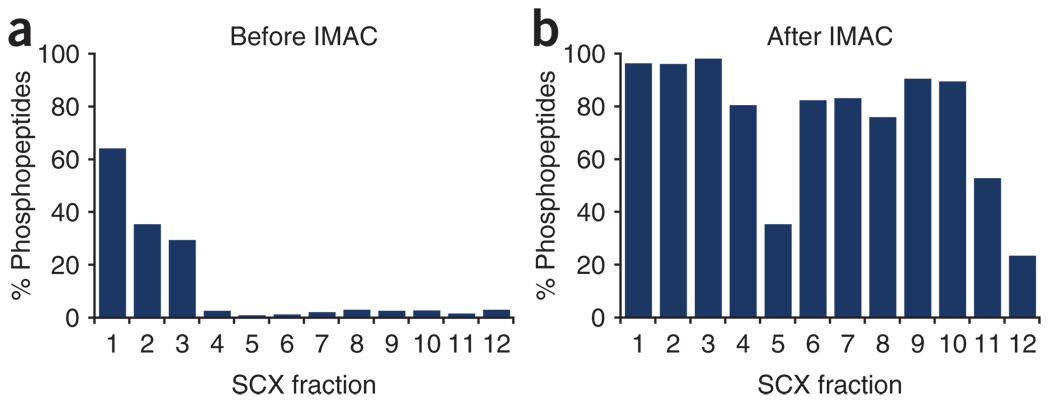
To show the phosphopeptide enrichment obtained in each step, the fraction of unique phosphopeptides over the total number of unique peptides identified within each fraction is plotted in Figure 3. Figure 3a shows the result after SCX chromatography only, and Figure 3b shows the result after both SCX chromatography and IMAC. Although SCX chromatography alone provides some phosphopeptide enrichment in the three earliest fractions13, coupling SCX chromatography with IMAC increases phosphopeptide enrichment to >75% for most fractions.
Figure 3. Enrichment obtained after each step represented as the fraction of phosphopeptides over all the peptides identified.
(a) SCX chromatography alone enriches for phosphopeptides in early fractions. (b) Further IMAC enrichment over SCX fractions produces >75% phosphopeptides in most samples.
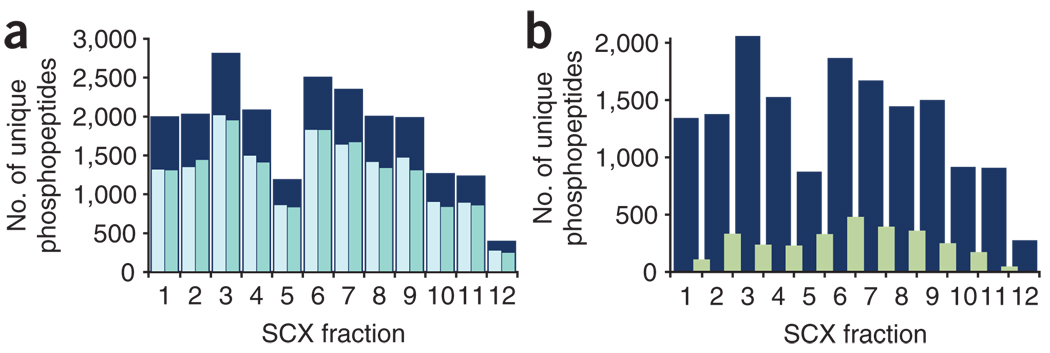
We have observed that running technical duplicates (i.e., analyzing the same sample in the mass spectrometer twice) of complex peptide or phosphopeptide mixtures significantly increases the number of unique identifications19,25, as the overlap between them is 70–75% (Fig. 4a). On the other hand, the number of phosphopeptides identified repeatedly in two consecutive fractions represents only 20% of the total phosphopeptides from either of the two fractions compared, revealing good resolution from the SCX chromatography (Fig. 4b).
Figure 4. Phosphopeptide distributions.
(a) Duplicate analyses of each fraction (pale blue and green) significantly increases the number of unique phosphopeptides identified (dark blue). (b) Overlap between consecutive SCX fractions (green) as compared with the number of unique phosphopeptides identified from one replicate of each fraction (dark blue).
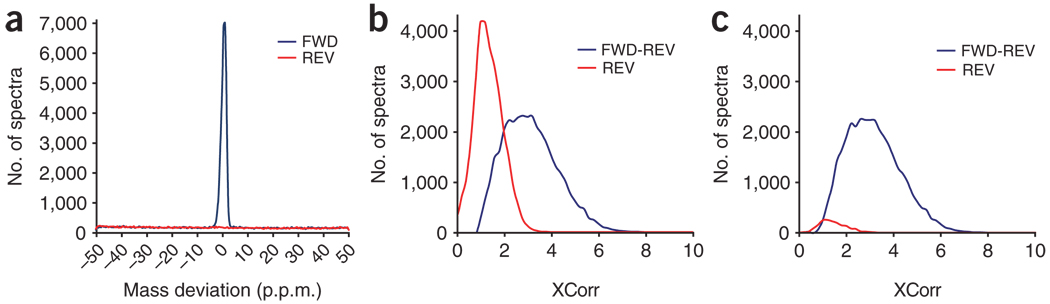
Finally, discrimination between right and wrong answers using Sequest XCorr score filtering alone would set the threshold at a very high value if a 1% false-discovery identification rate (FDR) is desired, causing a significant number of right but low scoring answers to be thrown away (Fig. 5a; red, reverse/wrong answers; blue, forward minus reverse/right answers). Measuring accurate precursor masses and performing searches in a mass tolerance window ~10 times bigger than the mass deviations measured (Fig. 5b) allows for an additional and powerful filtering criterion: mass error tolerance in a p.p.m. window25,29,30 that rescues many low XCorr scoring answers, while maintaining an ~1% FDR (Fig. 5c).
Figure 5. Contribution of mass accuracy to phosphopeptide identification rates.
(a) Distribution of mass deviations for all searched spectra. Matches to the forward database (blue) and those matching to the reverse database (red) are shown binned by 0.2 p.p.m. By searching with a larger precursor ion mass tolerance window than the distribution of true positives, many incorrect matches, which are uniformly distributed throughout the mass deviation window, can be removed. (b) Distribution of XCorr values of all searched spectra binned in 0.2 XCorr units. Matches to reversed sequences are shown in red. This represents one-half of all false-positives. Matches to the forward database with the number of reverse hits subtracted (true-positives) are shown in blue. (c) Same distribution as in b allowing only matches with a mass deviation between −2.5 to +2.5 p.p.m. By utilizing the known mass accuracy, most incorrect matches (in red) are removed from the data set. Additional filtering parameters such as dCn′ and solution charge can be used also to reduce the FDR; they will accept many correct answers with relatively lower XCorr values.
Data analysis
When generating large data sets, special attention should be paid to estimating, minimizing and reporting error rates of peptide identifications, to minimize erroneous annotations in phosphorylation databases. To this end, we introduced the target-decoy database-searching strategy28, where searches are performed against a single database that contains all proteins from the organism of interest, first in the forward and then in the reverse direction, and an FDR is estimated on the basis of the number of hits derived from flagged reverse proteins. Importantly, reverse matches can be used to guide threshold selection for mass deviation, XCorr, solution charge state and dCn′ parameters. Most laboratories performing large-scale phosphorylation studies have now adopted this simple and powerful approach for error rate determination20,31,32.
In addition to the issue of peptide false-discovery rates, analysis of phosphorylation (or other posttranslational modifications) by MS may introduce ambiguity associated with the precise position of the modification site. When more than one potential acceptor site exists in a peptide, some method of post-analysis verification should be used. The Ascore algorithm is a probability-based metric for assessing localization ambiguity29.
Besides these automated approaches for error-rate assessment and site localization, further validation for a subset of phosphopeptides might be desired, especially before follow-up biological experiments. Peptide synthesis constitutes the gold standard, where the MS/MS spectrum from the synthetic peptide is compared with the one acquired from the biological sample33. In addition, reanalysis of the sample using high-resolution MS/MS, different fragmentation techniques such as electron capture dissociation (ECD)34 and electron transfer dissociation (ETD)35 or fragmentation schemes such as neutral loss data-dependent MS3 (ref. 13) and multistage activation36 frequently bring additional confirmation for the phosphopeptide entity. Finally, computer-assisted manual inspection of MS/MS, looking for phosphopeptide diagnostic ions such as fragments derived from neutral loss of phosphoric acid, which often verify the presence of a phosphoserine or phosphothreonine in the sequence, and proline-driven fragmentation, has been regularly practiced13,16 with the drawback of being highly subjective, and correct discrimination is very much dependent on user expertise.
To perform database searches and data analysis, in our studies, RAW files were converted to the mzXML file format and imported into a MySQL relational database. MS/MS spectra were searched against a target-decoy28 S. cerevisiae ORFs database using the Sequest algorithm (version 27, revision 12), with 50 p.p.m. precursor mass tolerance, tryptic enzyme specificity with two missed cleavages allowed and static modification of cysteines (+57.02146, carboxamidomethylation). Dynamic modifications were 79.96633 Da on Ser, Thr and Tyr (phosphorylation) and 15.99491 Da on Met (oxidation). A maximum of four modifications of any one type and five total modifications were allowed per peptide. Sequest XCorr and dCn′18 score cutoffs were empirically determined for the entire data set, and mass deviation (in p.p.m.) and peptide solution charge filters were determined for each sample individually, using decoy matches as a guide28 and aiming to maximize the number of peptide spectral matches, although maintaining an estimated FDR of ≤ 1%. Identified phosphopeptides passing our filtering criteria were submitted to the Ascore algorithm29 for precise site localization.
ACKNOWLEDGMENTS
We thank Andrew Alpert from PolyLC for kindly providing columns for SCX chromatography and Manuel Rodriguez-Falcon for the initial tests of the combined IMAC-desalting procedure. We are also grateful to Joshua T. Wilson-Grady for constructive comments on the manuscript. This work was supported by NIH grant HG3456 to S.P.G.
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Hastie CJ, McLauchlan HJ, Cohen P. Assay of protein kinases using radiolabeled ATP: a protocol. Nat. Protoc. 2006;1:968–971. doi: 10.1038/nprot.2006.149. [DOI] [PubMed] [Google Scholar]
- 2.Collins MO, Yu L, Choudhary JS. Analysis of protein phosphorylation on a proteome-scale. Proteomics. 2007;7:2751–2768. doi: 10.1002/pmic.200700145. [DOI] [PubMed] [Google Scholar]
- 3.Hunter T, Cooper JA. Protein-tyrosine kinases. Annu. Rev. Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- 4.Rush J, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 5.Andersson L, Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal. Biochem. 1986;154:250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 6.Posewitz MC, Tempst P. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal. Chem. 1999;71:2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 7.Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 8.Sano A, Nakamura H. Titania as a chemo-affinity support for the column-switching HPLC analysis of phosphopeptides: application to the characterization of phosphorylation sites in proteins by combination with protease digestion and electrospray ionization mass spectrometry. Anal. Sci. 2004;20:861–864. doi: 10.2116/analsci.20.861. [DOI] [PubMed] [Google Scholar]
- 9.Kweon HK, Hakansson K. Selective zirconium dioxide-based enrichment of phosphorylated peptides for mass spectrometric analysis. Anal. Chem. 2006;78:1743–1749. doi: 10.1021/ac0522355. [DOI] [PubMed] [Google Scholar]
- 10.Holmes CF. A new method for the selective isolation of phosphoserine-containing peptides. FEBS Lett. 1987;215:21–24. doi: 10.1016/0014-5793(87)80106-0. [DOI] [PubMed] [Google Scholar]
- 11.Oda Y, Nagasu T, Chait BT. Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat. Biotechnol. 2001;19:379–382. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]
- 12.Zhou H, Watts JD, Aebersold R. A systematic approach to the analysis of protein phosphorylation. Nat. Biotechnol. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 13.Beausoleil SA, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, et al. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 15.Wilson-Grady JT, Villen J, Gygi SP. Phosphoproteome analysis of fission yeast. J. Proteome Res. 2008;7:1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- 16.Gruhler A, et al. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Trinidad JC, Specht CG, Thalhammer A, Schoepfer R, Burlingame AL. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteomics. 2006;5:914–922. doi: 10.1074/mcp.T500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. USA. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhai B, Villen J, Beausoleil SA, Mintseris J, Gygi SP. Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 2008;7:1675–1682. doi: 10.1021/pr700696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 21.Pinkse MW, et al. Highly robust, automated, and sensitive online TiO2-based phosphoproteomics applied to study endogenous phosphorylation in Drosophila melanogaster. J. Proteome Res. 2008;7:687–697. doi: 10.1021/pr700605z. [DOI] [PubMed] [Google Scholar]
- 22.Wu J, Shakey Q, Liu W, Schuller A, Follettie MT. Global profiling of phosphopeptides by titania affinity enrichment. J. Proteome Res. 2007;6:4684–4689. doi: 10.1021/pr070481m. [DOI] [PubMed] [Google Scholar]
- 23.Mazanek M, et al. Titanium dioxide as a chemo-affinity solid phase in offline phosphopeptide chromatography prior to HPLC-MS/MS analysis. Nat. Protoc. 2007;2:1059–1069. doi: 10.1038/nprot.2006.280. [DOI] [PubMed] [Google Scholar]
- 24.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protoc. 2006;1:1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 25.Bakalarski CE, Haas W, Dephoure NE, Gygi SP. The effects of mass accuracy, data acquisition speed, and search algorithm choice on peptide identification rates in phosphoproteomics. Anal. Bioanal. Chem. 2007;389:1409–1419. doi: 10.1007/s00216-007-1563-x. [DOI] [PubMed] [Google Scholar]
- 26.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 27.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 28.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 29.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 30.Haas W, et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Albuquerque CP, et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bodenmiller B, et al. PhosphoPep—a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol. Syst. Biol. 2007;3:139. doi: 10.1038/msb4100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zarling AL, et al. Phosphorylated peptides are naturally processed and presented by major histocompatibility complex class I molecules in vivo. J. Exp. Med. 2000;192:1755–1762. doi: 10.1084/jem.192.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stensballe A, Jensen ON, Olsen JV, Haselmann KF, Zubarev RA. Electron capture dissociation of singly and multiply phosphorylated peptides. Rapid Commun. Mass Spectrom. 2000;14:1793–1800. doi: 10.1002/1097-0231(20001015)14:19<1793::AID-RCM95>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 35.Chi A, et al. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. USA. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 2004;76:3590–3598. doi: 10.1021/ac0497104. [DOI] [PubMed] [Google Scholar]