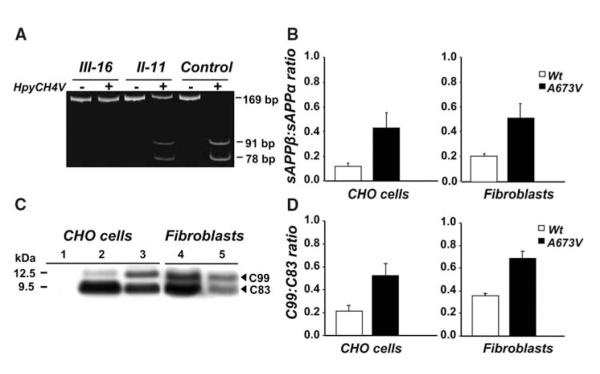
Fig. 1.
Analysis of APP gene and APP processing. (A) APP gene analysis by restriction fragment length polymorphism of 169—base pair (bp) polymerase chain reaction (PCR) products amplified from homozygous (III-16), heterozygous (II-11), and control subjects. In the absence of the A673V mutation, the enzyme HpyCH4V generates two fragments of 91 and 78 bp. The mutation abolishes the restriction site, and the PCR product remains uncut. (B) sAPPβ:sAPPα ratio in conditioned media from CHO cells transfected with wild-type or A673V-mutated APP and fibroblasts of the proband and four controls. Error bars represent means ± SD. (C) APP carboxy-terminal fragments C99 and C83 (arrowheads) in CHO cells transfected with wild-type (lane 2) or A673V-mutated (lane 3) APP and fibroblasts from a control (lane 4) and the proband (lane 5), as shown by immunoblot analysis. Lane 1 corresponds to from nontransfected CHO cells. (D) Densitometric analysis of immunoblots, showing a significant increase in the C99:C83 ratio (P = 0.0001) in cells carrying the A673V mutation. Error bars represent means ± SD.