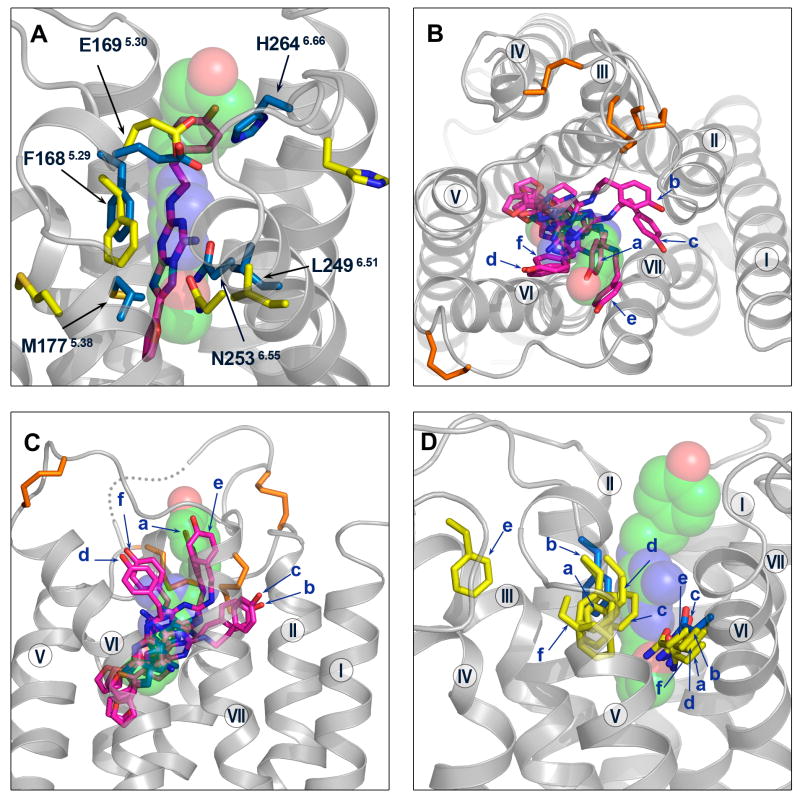
Figure 6.
Comparison between the best models and the crystal structure around the ligand-binding site. (A) The ligand and the ligand-binding residues F1685.29, E1695.30, M1775.38, L2496.51, N2536.55, H2646.66 are shown for the best model (Costanzi) and the crystal structure. The ligand is shown as sticks for the model (magenta colored carbon atoms) and semitransparent spheres (green colored carbon atoms) for the crystal structure; the ligand-binding residues are shown as yellow sticks for the model, and blue sticks for the crystal structure. Extracellular (B) and side views (C) of the ligand in the binding pocket for the best predictions from the top six groups (magenta sticks for models, and green spheres for the crystal structure). The receptor crystal structure is in gray ribbons. The disulfide bonds are shown in orange sticks (D) The ligand-binding residues F1685.29 and N2536.55 are shown as sticks for the best predictions from the top six groups (yellow for models, and blue for the crystal structure). In (B), (C) and (D) the models are labeled as a – Constanzi; b - Katritch; c - Lam; d - Davis; e - Maigret; f – Jurkowski.