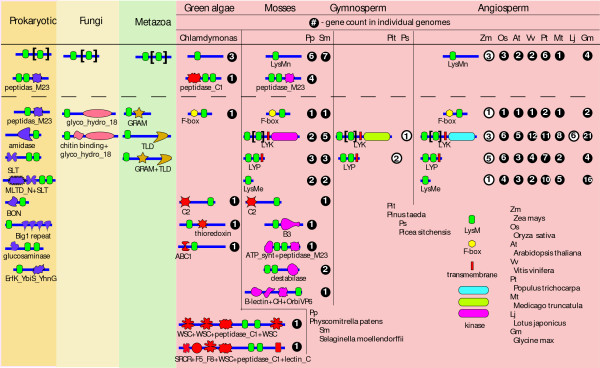
Figure 1.
The domain catalogue of LysM proteins across kingdoms. Only domain architectures with more than 5 occurrences in the Pfam database were diagramed roughly to scale. Domains were represented as different symbols and domain names were labeled beneath the diagrams except for the LysM domain, which is not labeled. The domain architectures above the dash line are conserved across different kingdoms and those below the dash line are distinct to each kingdom. The black brackets denote that the numbers of LysM are variable, ranging from 0 to 11 in prokaryotes, fungi and animal kingdoms and ranging from 0 to 2 in plant LYK proteins. The apparent lack of architectural diversity in Pinus is most likely due to the incomplete genome sequence of this species. The predicted subcellular localizations of LysM proteins in plants are also illustrated by drawing the transmembrane domain and drawing the diagram from N- to C-terminus. The incidence of individual architectures in plants is labeled on the right of the diagram. The numbers in filled circles denote the overall architectures in the entire genome and those in open circles are incomplete due to the lack of sufficient genome sequences.