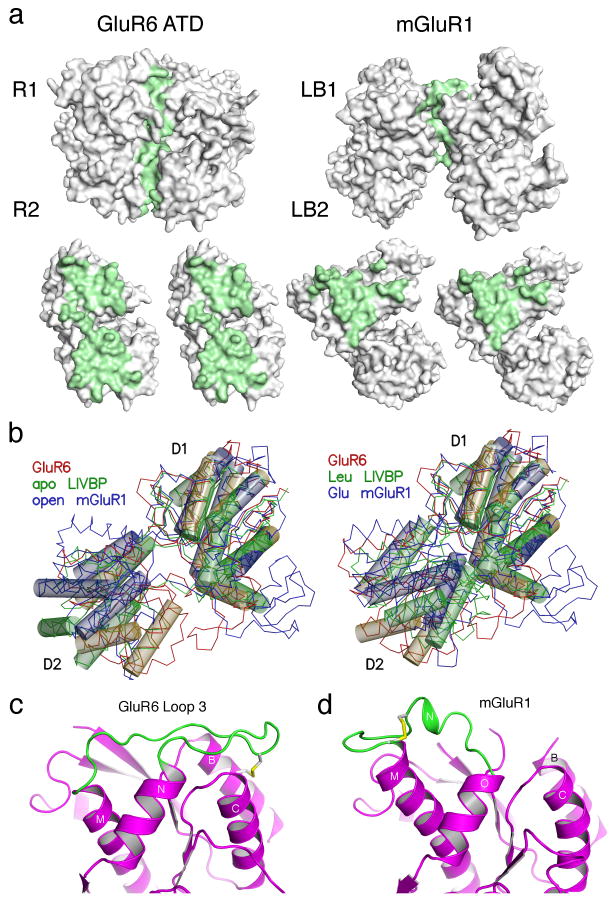
Figure 4.
The GluR6 ATD and mGluR dimers have different conformations. (a) Molecular surfaces for the GluR6 ATD and mGluR1 dimers crystallized in the ligand free open-open/R conformation (1EWT) for the dimer assembly, and after rotation by ± 90° of the partner subunits; residues with solvent inaccessible atoms are colored green. (b) The left panel shows crystal structures for the apo forms of LIVBP and mGluR1 superimposed on a GluR6 ATD protomer. Thin lines show Cα traces and transparent cylinders illustrate conserved α-helices which overlap when either the R1 or R2 domains are used for superposition. The right panel shows the superposition for the leucine and glutamate bound complexes of LIVBP and mGluR1. (c) Ribbon diagram showing the conformation of loop 3 and associated disulfide bridges in GluR6 ATD; the view is the same as in (a) and shows the left subunit in a dimer assembly. (d) The disulfide bridge and loop conformation in the corresponding region of mGluR1.