Figure 1. Unrooted phylogenetic tree of VP4 coding sequence.
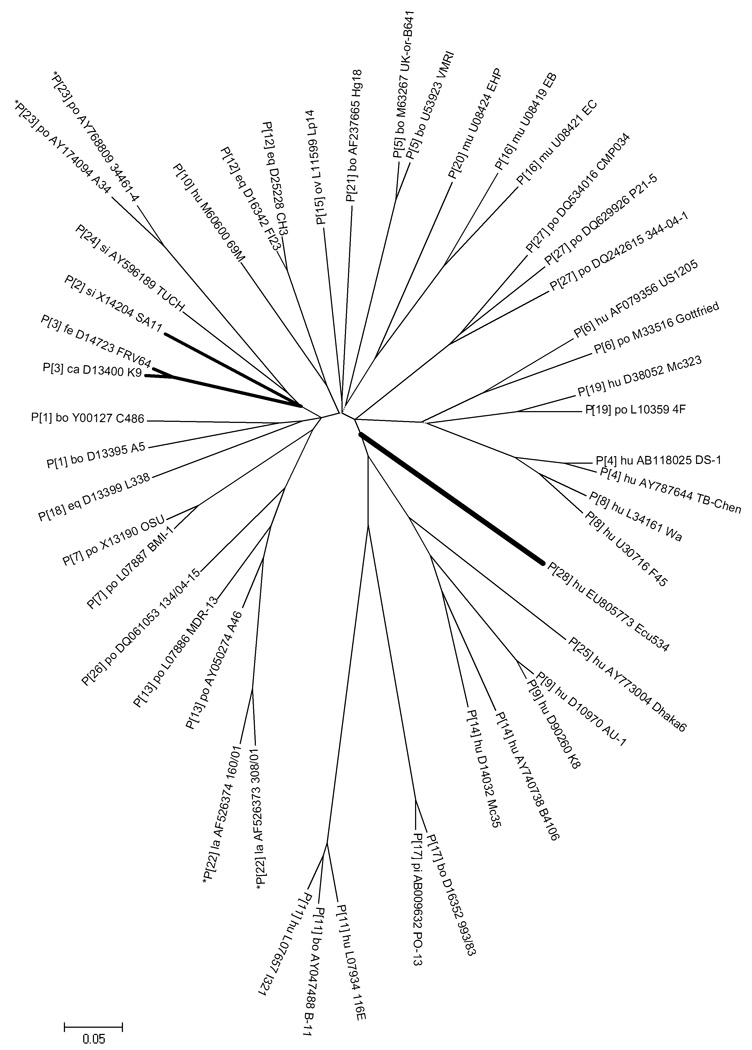
For each genotype, several representative sequences from GenBank are shown to provide perspective on the relative distance within and between genotypes. (The sequence alignment is available from the authors by request.) Strain Ecu534 (genotype P[28]) and three top BLAST hits are highlighted with bold branches. Evolutionary distances are in the units of base substitutions per site and were computed using the Maximum Composite Likelihood method (Tamura et al., 2004), as implemented by MEGA4 (Tamura et al., 2007). The optimal tree was constructed using the Neighbor-Joining method (Saitou and Nei, 1987). Nucleotide positions with missing data or gaps were eliminated only in pairwise sequence comparisons, which mainly affected the incomplete P[22] and P[23] genotype sequences (indicated by asterisks). All nodes have ≥90% bootstrap support, except for the three very short internal branches indicated by dotted lines.
