SUMMARY
Molecular aspects of the circadian clock in the cyanobacterium Synechococcus elongatus have been described in great detail. Three-dimensional structures have been determined for the three proteins, KaiA, KaiB, and KaiC, that comprise a central oscillator of the clock. Moreover, a temperature-compensated circadian rhythm of KaiC phosphorylation can be reconstituted in vitro with the addition of KaiA, KaiB, and ATP. These data suggest a relatively simple circadian system in which a single oscillator provides temporal information for all downstream processes. However, in vivo the situation is more complex, and additional components contribute to the maintenance of a normal period, the resetting of relative phases of circadian oscillations, and the control of rhythms of gene expression. We show here that two well-studied promoters in the S. elongatus genome report different circadian periods of expression under a given set of conditions in wild-type as well as mutant genetic backgrounds. Moreover, the period differs between these promoters with respect to modulation by light intensity, growth phase, and the presence or absence of a promoter-recognition subunit of RNA polymerase. These data contrast sharply with the current clock model in which a single Kai-based oscillator governs circadian period. Overall, these findings suggest that complex interactions between the circadian oscillator, perhaps other oscillators, and other cellular machinery result in a clock that is plastic and sensitive to the environment and to the physiological state of the cell.
Keywords: Circadian rhythms, cyanobacteria, circadian period, group 2 sigma factors
INTRODUCTION
Circadian rhythms of gene expression and physiological processes have been demonstrated in a wide spectrum of organisms, both prokaryotic and eukaryotic (Bell-Pedersen et al., 2005; Ditty et al., 2003). The cyanobacterium Synechococcus elongatus PCC 7942 has become the preeminent model organism for the study of prokaryotic circadian rhythms. S. elongatus cultures maintain a robust circadian period of gene expression, as assessed by luciferase reporter genes fused to cyanobacterial promoters (Kondo et al., 1993), even under conditions of rapid growth in which more than one generation is required to complete a circadian cycle (Kondo et al., 1997). The fidelity of both circadian period and relative phase is remarkably stable among progeny of a founder cell, without evidence of strong intercellular coupling (Mihalcescu et al., 2004). Three interacting proteins, KaiA, KaiB, and KaiC, comprise the only known S. elongatus circadian oscillator (Ishiura et al., 1998), and a circadian rhythm of KaiC phosphorylation can be reconstituted in vitro with only these three proteins and ATP (Nakajima et al., 2005). This in vitro oscillation is a dynamic dance of phosphorylation of KaiC driven by rhythmic interactions with KaiA and KaiB (Mori et al., 2007), which influence autokinase and dephosphorylation activities of KaiC. Data support a model in which KaiA enhances the autokinase activity of KaiC during subjective day (Circadian Time (CT) 0–12). Around CT12, KaiC becomes fully phosphorylated, which induces a conformation with higher affinity for KaiB. The KaiB-KaiC interaction results in an overall autophosphatase activity for KaiC, even for KaiA-KaiB–KaiC complexes. Once KaiC returns to its original unphosphorylated state, at CT24, its conformation changes back to one with low affinity for KaiB, and the interaction between KaiA and KaiC starts the circadian cycle all over again. Interchange of KaiC monomers keeps rhythmic phosphorylation of KaiC hexamers phase-coherent across the population, which results in a robust ensemble-averaged amplitude for several cycles (Mori et al., 2007).
The level of detail described for the in vitro reconstruction of the S. elongatus circadian oscillator suggests a simple self-supporting mechanism, but it is clear that the cellular clock is more complex. First, other proteins have been identified in vivo that interact with these oscillator components to provide input as modulators of free-running period and as cues for circadian entrainment, such that molecular oscillations are synchronized with environmental cycles (Ivleva et al., 2006; Smith & Williams, 2006; Takai et al., 2006). Secondly, clock output globally regulates expression of the entire genome, although promoters are expressed with different relative phasing, indicating specialization of circadian expression (Liu et al., 1995). The mechanism by which temporal information is transduced from the central Kai oscillator to the approximately 2,800 genes in the S. elongatus genome is not clear, but both chromosome compaction (Smith & Williams, 2006) and a signal transduction pathway that is closely associated with the oscillator (Iwasaki et al., 2000; Smith & Williams, 2006; Takai et al., 2006) are likely to globally support transcription rhythms.
Group 2 sigma factors, promoter-recognition subunits of bacterial RNA polymerase that are dispensable for growth under most conditions (Mulvey & Loewen, 1989), also affect both global and specialized patterns of circadian gene expression (Nair et al., 2002; Tsinoremas et al., 1996). Removing any one of the four group 2 sigma factors of S. elongatus that have been studied affects circadian properties of expression from a subset of tested promoters, suggesting that the combinatorial action of sigma factors contributes to wild-type (WT) circadian rhythmicity (Nair et al., 2002). It is especially striking that inactivation of one sigma factor gene, sigC, causes a lengthening of the period of expression from the psbAI promoter (PpsbAI) by about 2 h but does not affect that from the kaiBC and purF promoters (PkaiBC and PpurF, respectively) to the same extent, indicating that two or more oscillations of different periodicities can coexist in S. elongatus.
We recorded rhythms of bioluminescence from fusions of luciferase to the PpsbAI and PkaiBC in WT and sigC-null backgrounds for about three weeks, with the goal of verifying the presence of stable rhythms that run with different periods in S. elongatus. We found differences between the circadian periods of expression of these promoters in some conditions even in WT cells. The results showed that the circadian period of gene expression in S. elongatus can differ between loci with respect to their responses to differences in environmental light intensity (Aschoff, 1981), cell growth phase, and the absence of the SigC sigma factor. Overall, these findings suggest that complex interactions between two or more circadian oscillators and other cellular machinery result in a clock that is plastic and sensitive to the environment and the physiological state of the cell.
METHODS
Strains, Plasmids, and Growth Conditions
S. elongatus strains were grown under continuous light at 30 °C in liquid culture or on agar plates of modified BG-11 medium (BG-11M) (Bustos & Golden, 1991) with appropriate antibiotics (gentamycin 20 µg ml−1 and chloramphenicol 2 µg ml−1). Escherichia coli strain DH10B was used as the host for plasmids during cloning and it was manipulated by standard procedures (Sambrook et al., 1989).
Inactivation of sigma factor genes
Inactivation of the sigC gene has been described previously (Clerico et al., 2007; Nair et al., 2002). A gentamycin-resistance cassette was inserted in the open reading frame of the sigC gene in strain AMC669 (PpsbAI::luxAB and PpsbAI::luxCDE integrated in neutral site II [NS2, GenBank accession no. U44761]) to yield strain AMC1247, and in strain AMC1004 (PkaiB::luxAB in NS2 and PpsbAI::luxCDE in neutral site I [NS1, GenBank accession no. U30252]) to yield strain AMC1112. Transformants were selected on BG-11 M agar with gentamycin and chloramphenicol.
Bioluminescence assays
In addition to the luxAB gene set, all cyanobacterial strains used in this work carry a PpsbAI::luxCDE construct that directs synthesis in vivo of the long-chain aldehyde substrate for luciferase to make the reporter strains autonomously bioluminescent (Andersson et al., 2000). Bioluminescence was screened in a Packard TopCount luminometer (PerkinElmer Life Sciences) as previously described (Andersson et al., 2000; Mackey et al., 2007). Samples were inoculated onto BG–11 M agar in 96-well microtiter plates with proper antibiotics. The plates were incubated in constant light (LL) for 6–18 h, and then incubated in the dark for 12 h to reset the clocks of all of the cells in the population (Katayama et al., 1999). After resetting, the samples were incubated in LL and monitored in the luminometer for about 400 h, and the data were grouped into three time blocks: approximately the first 6 cycles (1–150 h), second 6 cycles (150–312 h), and remaining cycles of the run (312–400 h). Periods were calculated for 9–22 samples for each genotype and at least three independent runs were performed for each strain. In one set of experiments, after about 12 circadian cycles in LL, the samples were reset by incubation in the dark for 12 h and released in LL for 6 more cycles.
To control for the effect of light intensity on reporter gene expression levels during the TopCount run, comparisons were made between strains only from wells exposed to similar light intensities (Katayama et al., 2003), here designated as “low-light” or “high-light” intensity (approximately 25 or 100 micromoles photons × m−2 × s−1, respectively).
TopCount data were imported into Microsoft Excel 2000 using the Import & Analysis software package (S. A. Kay Laboratory, The Scripps Research Institute, La Jolla, CA, USA; (Plautz et al., 1997)), and phase, amplitude, and period were calculated by using the Fourier analysis software, FFT-NLLS, version 98.5, from the Import & Analysis software package (M. Straume, National Science Foundation Center for Biological Timing, University of Virginia). Some cycle-to-cycle variations in amplitude and levels of bioluminescence are normally observed in all the strains manipulated in the lab.
Statistical analysis of the data
Periods from long runs were calculated for the entire run or 6-cycle segments of the run using FFT-NLLS. Sets of period data were analyzed using Sigmastat 2.0 software (Systat Software). The Kruskal–Wallis One-Way Analysis of Variance on Ranks was used to determine whether or not the variances of the tested groups are statistically different, using P ≤ 0.05 as the significance criterion.
RESULTS AND DISCUSSION
In order to determine whether S. elongatus cells can stably maintain two circadian rhythms that have distinctly different periods, we recorded rhythms from PpsbAI and PkaiBC reporters in WT and sigC null backgrounds for 2–3 weeks under free-running conditions. The results from this single large dataset are reported here as selected individual bioluminescence traces (Fig. 1), a visual display of circadian periods from different treatment groups and strains (Fig. 2), and as a tabulation of statistical data (Table 1).
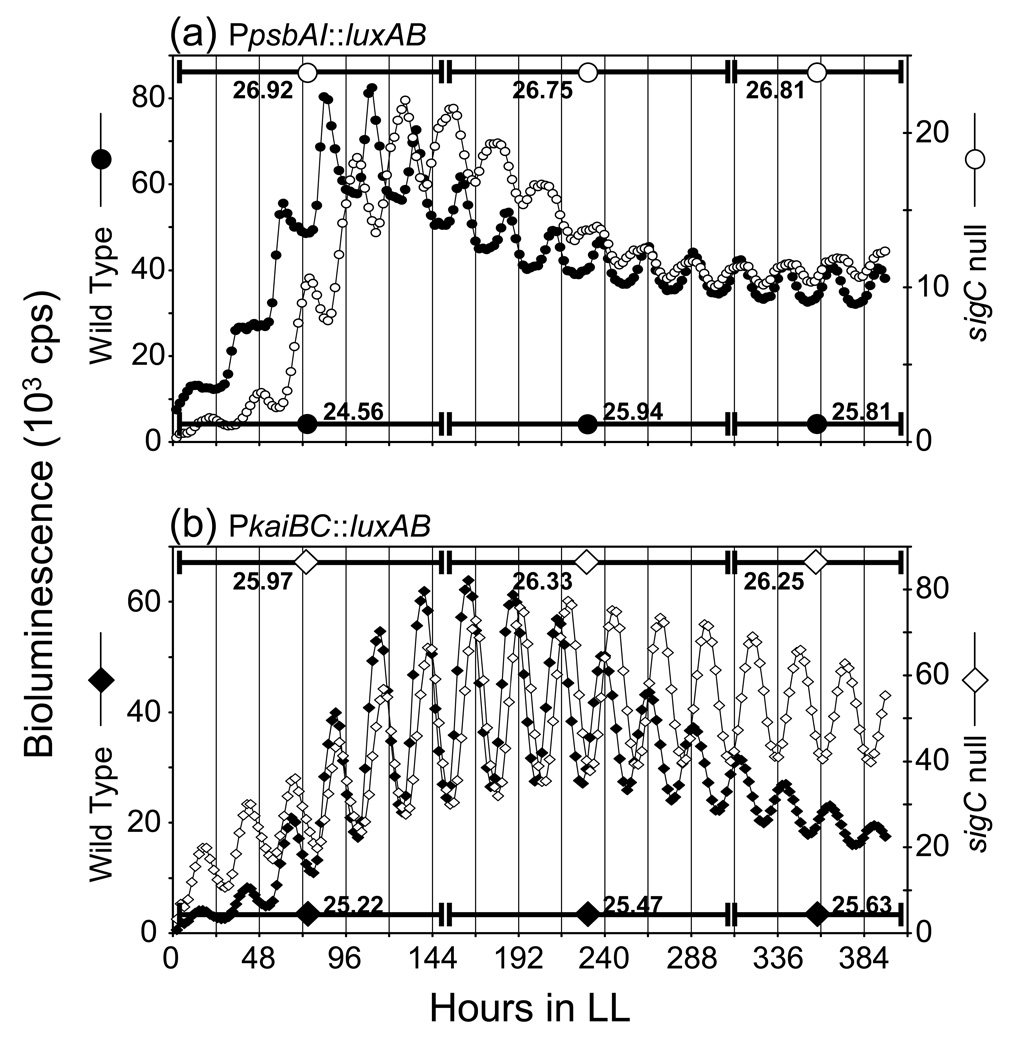
Figure 1.
Bioluminescence traces for PpsbAI::luxAB and PkaiBC::luxAB reporter strains. One representative trace for each strain is plotted. Bioluminescence (plotted as 103 counts per second) over time in h from samples monitored in constant low light from WT (closed symbols) and sigC-null strains (open symbols) that express (a) PpsbAI::luxAB reporter (circles); (b) PkaiBC::luxAB reporter (diamonds). The calculated periods (in h) for each portion of the curve are indicated for each strain at the bottom or the top of each plot. Absolute values of bioluminescence are related to cell number and do not provide a direct comparison of promoter strength; statistically identical data were obtained with different inocula of cells.
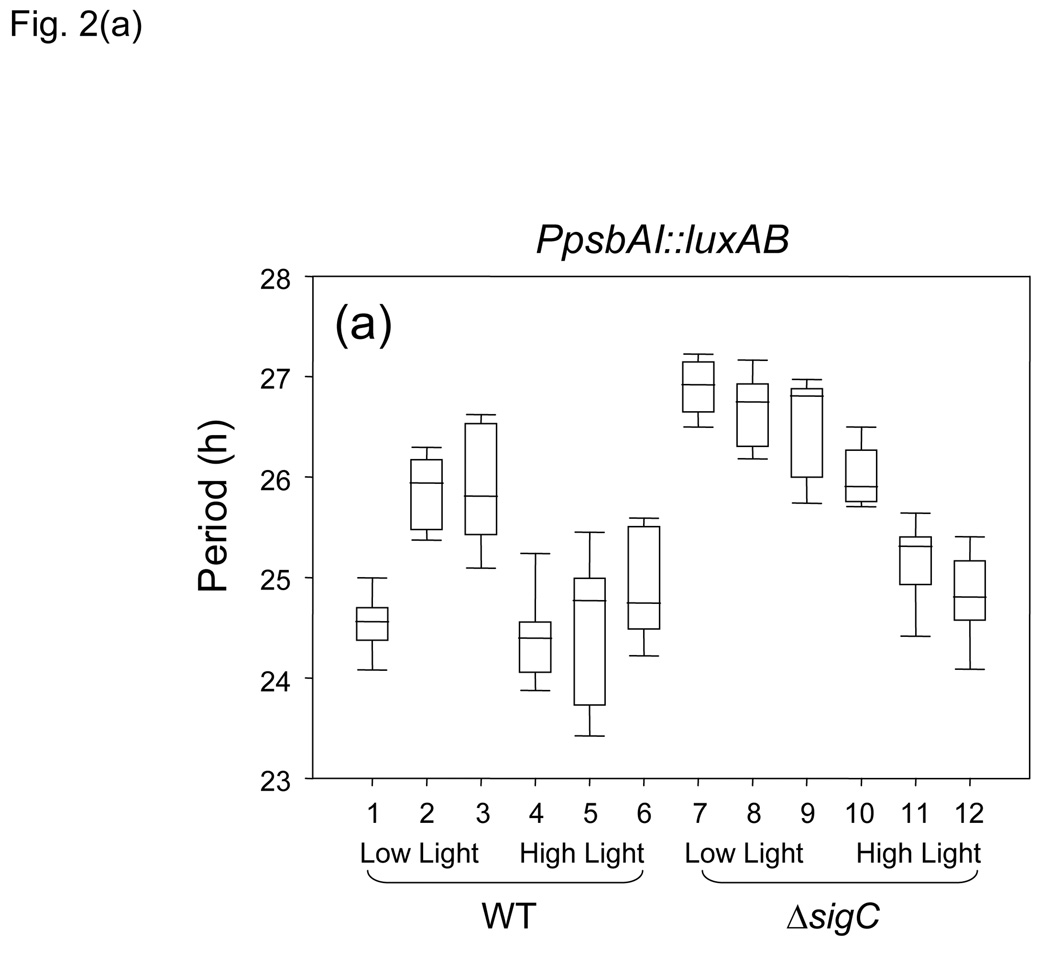
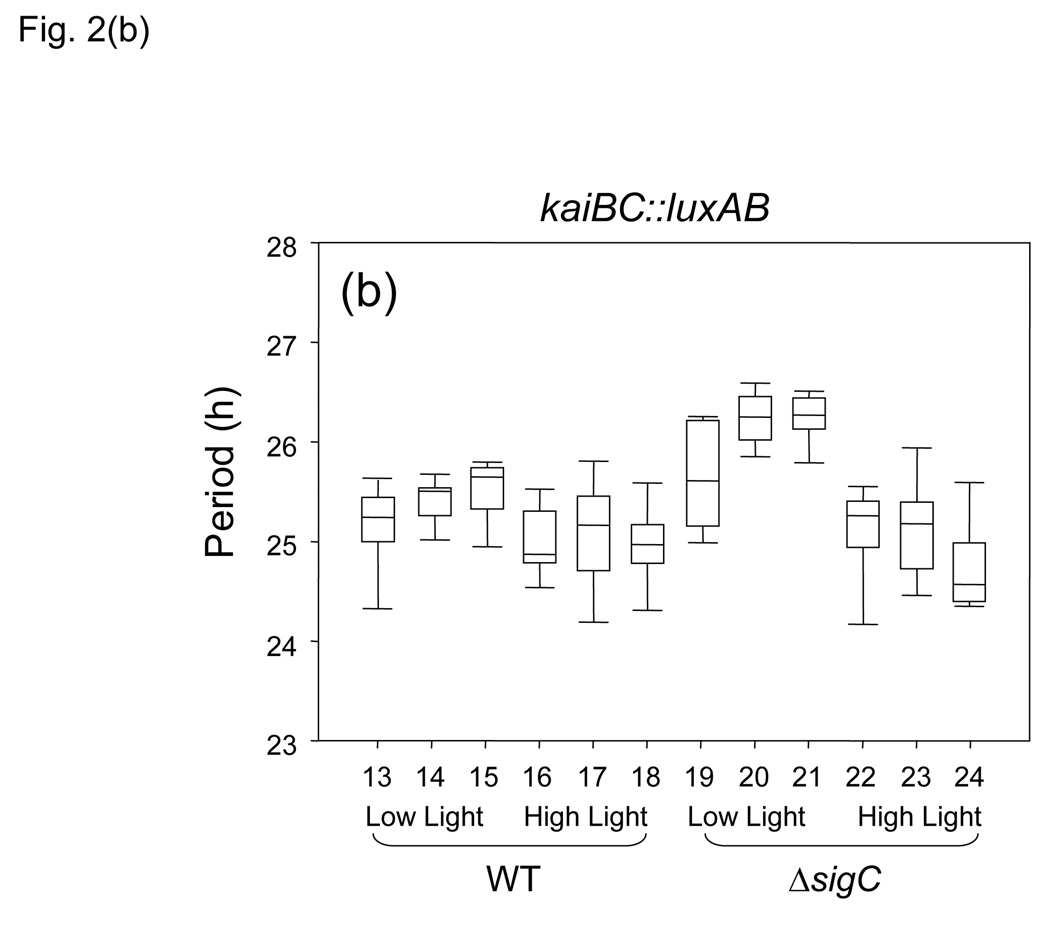
Figure 2.
Graphical representation of circadian periods from different reporter genes in specific environmental conditions and genetic backgrounds. Statistical analysis used Kruskal–Wallis One-Way Analysis of Variance on Ranks. Data are represented as boxplots, in which period is indicated by the vertical axis and each number on the horizontal axis designates a strain listed in Table 1. Each box is bisected by the median value for period, with whiskers representing the 5th and 95th percentiles of values. (a) PpsbAI::luxAB reporter; (b) PkaiBC::luxAB reporter; WT background samples 1–6 and 13–18; sigC null samples 7–12 and 19–24; low-light samples 1–3, 7–9, 13–15, and 19–21; high-light samples 4–6, 10–12, 16–18, and 22–24. The bin of hours during the 312–h sampling period that is indicated for each boxplot, also listed in Table 1, is: plots 1, 4, 7, 10, 13, 16, 19, and 22, 2–150 h; plots 2, 5, 8, 11, 14, 17, 20, and 23, 150–312; plots 3, 6, 9, 12, 15, 18, 21, and 24, 312–395 h.
Table 1.
| * | Reporter Gene | sigC | Light | Hours† | N‡ | Median§ | 25%§ | 75%§ |
|---|---|---|---|---|---|---|---|---|
| 1 | PpsbAI::luxAB | WT | Low | 2–150 | 13 | 24.56 | 24.37 | 24.68 |
| 2 | PpsbAI::luxAB | WT | Low | 150–312 | 17 | 25.94 | 25.48 | 26.15 |
| 3 | PpsbAI::luxAB | WT | Low | 312–395 | 17 | 25.81 | 25.45 | 26.52 |
| 4 | PpsbAI::luxAB | WT | High | 2–150 | 14 | 24.39 | 24.06 | 24.52 |
| 5 | PpsbAI::luxAB | WT | High | 150–312 | 14 | 24.77 | 23.79 | 24.99 |
| 6 | PpsbAI::luxAB | WT | High | 312–395 | 14 | 24.74 | 24.51 | 25.51 |
| 7 | PpsbAI::luxAB | Null | Low | 2–150 | 13 | 26.92 | 26.67 | 27.13 |
| 8 | PpsbAI::luxAB | Null | Low | 150–312 | 13 | 26.75 | 26.30 | 26.92 |
| 9 | PpsbAI::luxAB | Null | Low | 312–395 | 13 | 26.81 | 26.05 | 26.86 |
| 10 | PpsbAI::luxAB | Null | High | 2–150 | 14 | 25.90 | 25.76 | 26.27 |
| 11 | PpsbAI::luxAB | Null | High | 150–312 | 14 | 25.31 | 25.03 | 25.40 |
| 12 | PpsbAI::luxAB | Null | High | 312–395 | 14 | 24.80 | 24.59 | 25.16 |
| 13 | PkaiBC::luxAB | WT | Low | 2–150 | 22 | 25.22 | 25.01 | 25.41 |
| 14 | PkaiBC::luxAB | WT | Low | 150–312 | 22 | 25.47 | 25.55 | 25.30 |
| 15 | PkaiBC::luxAB | WT | Low | 312–395 | 17 | 25.63 | 25.33 | 25.72 |
| 16 | PkaiBC::luxAB | WT | High | 2–150 | 21 | 24.85 | 24.77 | 25.25 |
| 17 | PkaiBC::luxAB | WT | High | 150–312 | 22 | 25.14 | 24.70 | 25.42 |
| 18 | PkaiBC::luxAB | WT | High | 312–395 | 19 | 24.95 | 24.77 | 25.14 |
| 19 | PkaiBC::luxAB | Null | Low | 2–150 | 13 | 25.59 | 25.13 | 26.19 |
| 20 | PkaiBC::luxAB | Null | Low | 150–312 | 13 | 26.23 | 26.00 | 26.42 |
| 21 | PkaiBC::luxAB | Null | Low | 312–395 | 9 | 26.25 | 26.14 | 26.41 |
| 22 | PkaiBC::luxAB | Null | High | 2–150 | 9 | 25.24 | 24.92 | 25.38 |
| 23 | PkaiBC::luxAB | Null | High | 150–312 | 9 | 25.16 | 24.75 | 25.30 |
| 24 | PkaiBC::luxAB | Null | High | 312–395 | 10 | 24.55 | 24.39 | 24.93 |
Numbers refer to boxplots in Fig. 2
Hours range indicates the portion of the 395-h monitoring interval that is included in each set of data
Individual samples measured, each of which includes approximately 6 cycles in the period calculation; N includes samples from at least 3 independent monitoring experiments
median and lower and upper quartiles of period values from Kruskal–Wallis One-Way Analysis of Variance on Ranks
Based on previous observations (Nair et al., 2002), we predicted that, over time, the PpsbAI WT bioluminescence peak would fall increasingly distant from that of the sigC mutant, whose circadian period is 2 h longer, and that after several days the traces would appear to cross, with a full additional cycle present in the WT relative to the sigC mutant. As predicted, after 150 h the PpsbAI traces from the two genetic backgrounds, monitored at low constant light levels, were almost antiphase, with the WT strain having completed six cycles and the mutant approximately five and one-half (Fig. 1a). However, during the next 245 h, after the two traces came back in phase, their phase relationship did not change appreciably, indicating that a period change had occurred in one or both strains. We calculated the periods for different portions of the experimental run for each strain. The results show that, under low light (Table 1, rows 1–3 and 7–9, and Fig. 2a), after about six circadian cycles the period reported by PpsbAI in the WT background lengthened by about 1.5 h and became stable for the duration of monitoring. The period length in the sigC mutant, which showed a 2 h longer period than WT in the first six cycles (compare lines 1 and 7 of Table 1, and Fig. 2a), did not change over time in the following six cycles (compare lines 7–9 of Table 1, and Fig. 2a), accounting for the apparent synchronization of the two traces after the first 150 h, when the difference in period between the two rhythms narrowed from 2.3 h to about 1.0 h (Fig. 1a, Fig. 2a, and Table 1). The comparisons shown in Fig. 1(a) derive from samples that were monitored under relatively low-light conditions; the period lengthening of PpsbAI expression in WT after six cycles was not observed in high-light conditions (Table 1, rows 4–6, and Fig. 2a).
Comparison of the circadian periods of PkaiBC reporter expression in WT and sigC mutant backgrounds in the first 150 h under these low-light monitoring conditions revealed no significant differences in period between the strains (Fig. 1b, Fig. 2b, and Table 1, rows 13–15 and 19–21), as reported previously (Nair et al., 2002).
Remarkably, this analysis showed a notable difference in the circadian periods of expression from the PpsbAI and PkaiBC reporters in WT cells under specific conditions of illumination. Direct comparison of the two traces in Fig. 1(c) shows that, at the beginning of the run in low-light conditions, PpsbAI-driven bioluminescence proceeds with a shorter period than that driven by PkaiBC (Table 1, rows 1 and 13); however, the phenomenon of period lengthening in later cycles from the PpsbAI reporter, which is not seen from the PkaiBC reporter, causes the two bioluminescence patterns to get back in phase around the seventh circadian cycle.
Additional analyses examined the period relationships of these genotypes under high-light conditions, which are known to have a period-shortening effect in WT cells (Aschoff, 1981; Katayama et al., 2003) that is more evident for PpsbAI than for PkaiBC (Table 1, rows 4–6 and 16–18, and Fig. 2a and Fig. 2b). In WT cells the period-lengthening of PpsbAI expression that occurs after six cycles at low light (Fig. 1a and Table 1, rows 1–3) was absent under high-light conditions (Fig. 2a and Table 1, rows 4–6). Moreover, the period in the sigC null strain shortened after 150 h (Fig. 2a and Table 1, rows 10–12). Thus, under high-light conditions after a week in free-running conditions, PpsbAI-driven bioluminescence cycles in the WT and sigC null backgrounds showed similar circadian periods (Fig. 2a and Table 1, rows 2–3 and 11–12).
No statistically significant differences were found between the periods reported by PkaiBC::luxAB in the WT strain versus the sigC mutant under high light (Fig. 2b and Table 1, rows 16–18 and 22–24). However, the values for the sigC null strain itself were significantly shorter under high-light than under low-light conditions after 150 h (Fig. 2b and Table 1, compare rows 20 and 23). The role of SigC has been investigated for another cyanobacterium, Synechocystis sp. strain PCC 6803 (Asayama et al., 2004). In that strain, the glnB nitrogen-regulated promoter is specifically recognized by SigC in stationary phase under conditions of nitrogen starvation, indicating that SigC exerts a function in stationary phase and is related with the control of metabolism under specific conditions. The lack of sigC may emulate a physiological condition typical of stationary phase that, in WT cells, occurs only after 150 h of growth. However, this explanation is insufficient to account for the effect of the sigC mutation on PkaiBC circadian expression, which acquires period shortening under high light that is not evident in WT cells.
We tested to see whether the period lengthening that is observed for PpsbAI expression after the first week of low-light monitoring is due to a change in entrainment properties of the clock or to a stable change in physiological state of the cell. We performed the experiment as shown in Fig. 1(a), but after 12 circadian cycles the cells were given a 12 h pulse of darkness to reset the clock and initiate a new free run (data not shown). When the samples emerged from darkness, they exhibited the longer period characteristic of late cycles (Figure 2a and Table 1, rows 2–3): 25.31±0.31 h for the six cycles before dark pulse and 25.56±0.09 h for the six cycles after the dark pulse. We concluded that the cells had entered a stable physiological state that lengthened circadian period irrespective of the elapsed interval since the last phase-setting cue. In the case of the PkaiBC reporter strain, a 12 h pulse of darkness did not have any effect on the circadian period of subsequent cycles.
This onset of PpsbAI period lengthening likely represents the point at which S. elongatus enters stationary growth phase. On agar plates, S. elongatus cells divide exponentially during microcolony formation and continue to divide to form multilayered colonies. Then, the growth rate slows down as result of light limitation for the cells in the interior of the colony (Kondo et al., 1997). In the present study, the configuration of the TopCount luminometer does not allow us to test for a change in growth rate of cells. An effect of metabolic state on the circadian clock has been shown before for cyanobacteria (Ivleva et al., 2005; Katayama et al., 2003). LdpA, a redox-sensitive protein that contains two Fe4S4 clusters, modulates the abundance of some components of the clock and tunes the length of the circadian period via signals that reflect the redox state of the cell. The current data suggest that the physiological state of the cell influences the clock in a gene-specific manner.
Consistent with previous analyses, we observed that the circadian rhythm of PpsbAI-driven luciferase bioluminescence is more sensitive to the effects of some mutations than is that from a PkaiBC reporter. Mutation of cpmA (circadian phase modifier) changes the relative timing of expression peaks from PpsbAI but not from PkaiBC (Katayama et al., 1999); loss of sasA renders PpsbAI nearly arrhythmic but leaves PkaiBC robustly rhythmic, although with a lower amplitude than in a WT background (Iwasaki et al., 2000); and inactivation of sigC lengthens the circadian period of expression from PpsbAI but not from PkaiBC (Nair et al., 2002).
Another outcome of this analysis was the finding that PkaiBC expression does not follow Aschoff’s Circadian Rule (Aschoff, 1981), which states that diurnal organisms exhibit slightly shorter circadian periods under higher light conditions than under lower light; such differences in period between high- and low-light samples in a given monitoring run are easily evident for expression of psbAI (Katayama et al., 2003). No differences in period of the PkaiBC rhythms were observed between high- and low-light samples. As a result, detection of a difference in circadian period between expression of PkaiBC and PpsbAI depends on both ambient light intensity and the number of cycles since inoculation of the monitoring plate that are used in the analysis.
The plasticity of circadian period also involves the group 2 sigma factors. In a sigC null strain, the PpsbAI reporter rhythm has a longer period than in the WT for the first several days. The fact that this phenomenon is not observed for the PKaiBC reporter suggests the presence of more than one oscillator operating in the same cell, as noted previously (Nair et al., 2002).
The concept of multiple oscillators within single-celled organisms is not without precedence. For example, the dinoflagellate Gonyaulax polyedra expresses circadian rhythms of multiple processes, including endogenous bioluminescence, bioluminescent flashing, photosynthesis, and aggregation that can be uncoupled under differential lighting or nutritional conditions (Roenneberg & Morse, 1993). These observations strongly suggest multiple oscillators underlying each of these behaviors that are coupled under most natural conditions. The selective advantage for this scenario may include lability in response properties to different nutritional, photic, and/or toxicological environments, enabling the “hands” of the underlying clocks to adapt rapidly to specific environmental cues without disproportionately altering the system as a whole. In this scenario, one might imagine that photosynthetic capacity, exemplified here by psbAI activity, may be more sensitive to environmental lighting than kaiBC activity, which comprises part of the central core oscillator.
Kitayama et al. recently reported that temperature-compensated circadian rhythms of gene expression can persist under conditions in which the oscillation of KaiC phosphorylation is blocked, consistent with a more complex system of oscillators in the cell (Kitayama et al., 2008). Nonetheless, to date, all known circadian oscillations in the cell, including the long-period psbAI rhythm in the sigC mutant, depend on the Kai oscillator (Nair et al., 2002). The tailored patterns of circadian activity exhibited by different promoters suggest that one or more additional timing circuits contributes to the intact clock of S. elongatus in vivo.
CONCLUSIONS
Extended monitoring of strains that report the activity of different promoters revealed that the psbAI and kaiBC promoters are expressed with different circadian periods under some situations even in WT cells, suggestive of more than one oscillator in the circadian clock. Differences in circadian period of PpsbAI activity relative to that of PkaiBC include: greater sensitivity of PpsbAI period to incident light intensity in wild-type backgrounds; acquisition of a light-sensitive period by PkaiBC when SigC is absent; period lengthening of PpsbAI activity after 6 days that was not observed for PkaiBC and which is lost when SigC is absent. All of the data indicate that multiple circadian periodicities can coexist in S. elongatus cells, consistent with a clock mechanism that is more complex than the phosphorylation cycle of KaiC, and that the physiological state of the cell influences the S. elongatus clock in a gene-specific fashion.
ACKNOWLEDGMENTS
We thank Dr. Shannon Mackey for critical reading of the manuscript and assistance with figures, Lily Bartoszek for proofreading, Dr. Jayna Ditty for her help in early stages of this work, and Jiffin Paulose for his help with Sigmastat software. This work was supported by a program project grant from the National Institutes of Health (P01 NS39546).
Abbreviations
- CT
circadian time
- NS1
Neutral Site I
- NS2
Neutral Site II
- WT
wild type
REFERENCES
- Andersson CR, Tsinoremas NF, Shelton J, Lebedeva NV, Yarrow J, Min H, Golden SS. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 2000;305:527–542. doi: 10.1016/s0076-6879(00)05511-7. [DOI] [PubMed] [Google Scholar]
- Asayama M, Imamura S, Yoshihara S, et al. SigC, the group 2 sigma factor of RNA polymerase, contributes to the late-stage gene expression and nitrogen promoter recognition in the cyanobacterium Synechocystis sp. strain PCC 6803. Biosci Biotechnol Biochem. 2004;68:477–487. doi: 10.1271/bbb.68.477. [DOI] [PubMed] [Google Scholar]
- Aschoff J. Freerunning and Entrained Circadian Rhythms. In: Aschoff J, editor. Handbook of Behavioral Neurobiology: Biological Rhythms. New York: Plenum Press; 1981. pp. 81–93. [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos SA, Golden SS. Expression of the psbDII gene in Synechococcus sp. strain PCC 7942 requires sequences downstream of the transcription start site. Journal of bacteriology. 1991;173:7525–7533. doi: 10.1128/jb.173.23.7525-7533.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerico EM, Ditty JL, Golden SS. Specialized techniques for site-directed mutagenesis in cyanobacteria. Methods Mol Biol. 2007;362:155–171. doi: 10.1007/978-1-59745-257-1_11. [DOI] [PubMed] [Google Scholar]
- Ditty JL, Williams SB, Golden SS. A cyanobacterial circadian timing mechanism. Annu Rev Genet. 2003;37:513–543. doi: 10.1146/annurev.genet.37.110801.142716. [DOI] [PubMed] [Google Scholar]
- Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: a component of the circadian clock senses redox state of the cell. Embo J. 2005;24:1202–1210. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva NB, Gao T, LiWang AC, Golden SS. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc Natl Acad Sci U S A. 2006;103:17468–17473. doi: 10.1073/pnas.0606639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Williams SB, Kitayama Y, Ishiura M, Golden SS, Kondo T. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- Katayama M, Tsinoremas NF, Kondo T, Golden SS. cpmA, a gene involved in an output pathway of the cyanobacterial circadian system. Journal of bacteriology. 1999;181:3516–3524. doi: 10.1128/jb.181.11.3516-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama M, Kondo T, Xiong J, Golden SS. ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942. Journal of bacteriology. 2003;185:1415–1422. doi: 10.1128/JB.185.4.1415-1422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama Y, Nishiwaki T, Terauchi K, Kondo T. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes & development. 2008;22:1513–1521. doi: 10.1101/gad.1661808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90:5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Mori T, Lebedeva NV, Aoki S, Ishiura M, Golden SS. Circadian rhythms in rapidly dividing cyanobacteria. Science. 1997;275:224–227. doi: 10.1126/science.275.5297.224. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M, Kondo T. Circadian orchestration of gene expression in cyanobacteria. Genes Devel. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- Mackey SR, Ditty JL, Clerico EM, Golden SS. Detection of rhythmic bioluminescence from luciferase reporters in cyanobacteria. Methods Mol Biol. 2007;362:115–129. doi: 10.1007/978-1-59745-257-1_8. [DOI] [PubMed] [Google Scholar]
- Mihalcescu I, Hsing W, Leibler S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature. 2004;430:81–85. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- Mori T, Williams DR, Byrne MO, Qin X, Egli M, McHaourab HS, Stewart PL, Johnson CH. Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol. 2007;5:e93. doi: 10.1371/journal.pbio.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey MR, Loewen PC. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel sigma transcription factor. Nucleic Acids Res. 1989;17:9979–9991. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Ditty JL, Min H, Golden SS. Roles for sigma factors in global circadian regulation of the cyanobacterial genome. Journal of bacteriology. 2002;184:3530–3538. doi: 10.1128/JB.184.13.3530-3538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Morse D. Two circadian oscillators in one cell. Nature. 1993;362:362–364. doi: 10.1038/362362a0. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Mittag M. The circadian program of algae. Seminars in Cell & Developmental Biology. 1996;7:753–763. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. NY: Cold Spring Harbor; 1989. [Google Scholar]
- Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai N, Nakajima M, Oyama T, Kito R, Sugita C, Sugita M, Kondo T, Iwasaki H. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci U S A. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsinoremas NF, Ishiura M, Kondo T, Andersson CR, Tanaka K, Takahashi H, Johnson CH, Golden SS. A sigma factor that modifies the circadian expression of a subset of genes in cyanobacteria. EMBO. 1996;15:2488–2495. [PMC free article] [PubMed] [Google Scholar]