Abstract
PKR (protein kinase R) is induced by interferon and is a key component of the innate immunity antiviral pathway. Upon binding dsRNA, PKR undergoes autophosphorylation reactions that activate the kinase leading it to phosphorylate eIF2α, thus inhibiting protein synthesis in virally-infected cells. PKR contains a dsRNA binding domain (dsRBD) and a kinase domain. The dsRBD is composed of two tandem dsRNA binding motifs. An autoinhibition model for PKR has been proposed whereby dsRNA binding activates the enzyme by inducing a conformational change that relieves the latent enzyme of the inhibition that is mediated by interaction of the dsRBD with the kinase. However, recent biophysical data support an open conformation for the latent enzyme where activation is mediated by dimerization of PKR induced upon binding dsRNA. We have probed the importance of interdomain contacts by comparing the relative stabilities of isolated domains with the same domain in the context of the intact enzyme using equilibrium chemical denaturation experiments. The two dsRNA binding motifs fold independently, with C-terminal motif exhibiting greater stability. The kinase domain is stabilized by about 1.5 kcal/mole in the context of the holenzyme and we detect low-affinity binding of the kinase and dsRBD constructs in solution, indicating that these domains interact weakly. Limited proteolysis measurements confirm the expected domain boundaries and reveal that the activation loop in the kinase is accessible to cleavage and unstructured. Autophosphorylation induces a conformation change that blocks proteolysis of the activation loop.
Protein kinase R (PKR) is an interferon-induced kinase that plays a key role in the innate immunity response to viral infection in higher eukaryotes (1–3). PKR has also been implicated in a variety of cellular signal transduction pathways (4, 5). The enzyme is synthesized in a latent state, but upon binding dsRNA, or structured RNAs containing dsRNA regions, it undergoes autophosphorylation at multiple serine and threonine residues resulting in activation. The most well characterized cellular substrate of PKR is the alpha subunit of initiation factor eIF2. Phosphorylation of eIF2α inhibits the initiation of translation. Thus, production of dsRNA that occurs during infection with viruses with positive-stranded RNA, dsRNA or DNA genomes (6) can result in PKR activation and subsequent inhibition of viral and host protein synthesis (7).
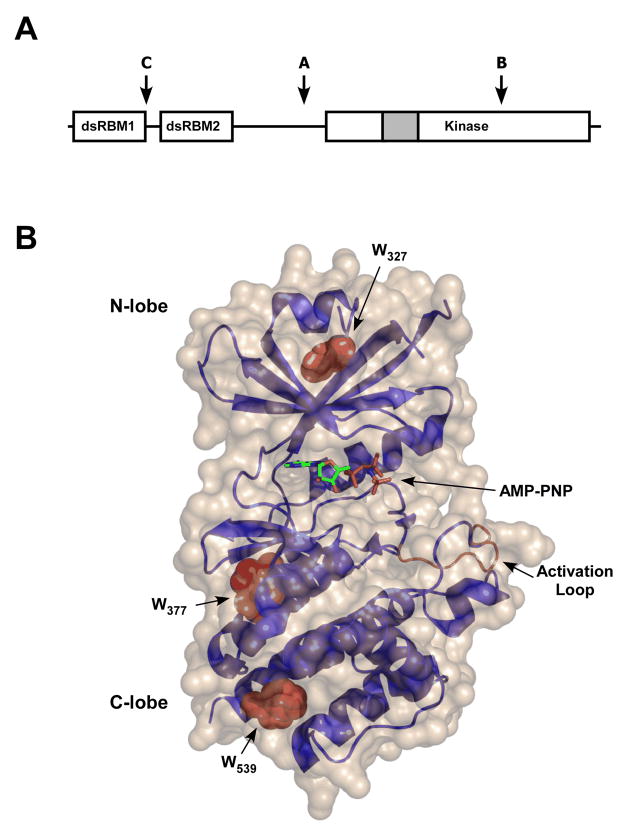
PKR is a 62 kDa protein that contains an N-terminal double-stranded RNA binding domain (dsRBD) and C-terminal kinase domain, with a central region of unknown function (Figure 1A). The dsRBD consists of two tandem copies of the ~70 amino-acid dsRNA binding motif (dsRBM) (8), dsRBM1 and dsRBM2. In the NMR structure of the PKR dsRBD, the two motifs each adopt the canonical αβββα dsRBM fold and are connected by an unstructured linker of ~ 20 amino acids (9). The overall structures of the two domains are quite similar, with a backbone RMSD of 2Å (9). However, dsRBM1 does exhibit significantly more backbone motion on the millisecond timescale than dsRBM2 (10). The X-ray structure of a complex of the PKR kinase domain with an N-terminal fragment of eIF2α was recently solved (11). Like other protein kinases (12), the catalytic domain of PKR contains two lobes: the N-terminal lobe is smaller and predominantly β sheet and the C-lobe is mostly helical (Figure 1B). The region between dsRBM2 and the kinase domain appears unstructured or dynamic in TROSY-HSQC spectra (13).
Figure 1.
PKR structure. A: Schematic representation of PKR domain organization. The letters “A,B,C” refer to the principal trypsin cleavage sites. The grey portion of the kinase domain corresponds to the kinase insert region. B: Structure of PKR kinase domain. The protein backbone is shown as a ribbon diagram in blue with the activation loop in brown. The three tryptophan side chains are shown in Van der Waals representation in red and AMPPNP is drawn in stick format. The solvent accessible surface is beige. The coordinates (PDB 2A19) were obtained from the crystal structure of a complex of the PKR kinase domain with eIF2α and AMPPNP (11) and the figure was rendered with PYMOL (Delano Scientific, Palo Alto, CA).
It is not well understood how RNA binding at the dsRBD results in activation of the kinase and several models have been proposed (14). In the autoinhibition model, latent PKR exist in a closed conformation in which the dsRBD interacts with the kinase domain and blocks substrate binding. RNA then functions to activate the kinase by binding to the dsRBD and inducing a conformational change that exposes the active site. In support of this mechanism, an interaction of dsRBM2 with the kinase domain has been reported based on NMR chemical shift perturbation experiments (10, 15). In contrast to this picture of a closed conformation for the latent enzyme, sedimentation velocity (16), AFM (17) and neutron scattering experiments (18, 19) suggest that PKR can adopt an extended structure in solution and nucleotide substrates have free access to the active site in both active and inactive forms of PKR (17). NMR resonances associated with the dsRBD are essentially superimposable in spectra of the isolated domain and full length enzyme (13), which argues against strong interactions of the dsRBD and kinase.
It has long been recognized that PKR is capable of dimerizing in the absence and the presence of dsRNA (16, 18, 20–22) and recent structural and biophysical data now favor models where dsRNA principally functions to enhance dimerization of PKR. The PKR kinase-eIF2α complex crystallized as a back-to-back dimer with the interface formed by the kinase N-terminal lobes (11). Helix αC in the N-lobe comprises part of the dimer interface and conformational changes in this helix often regulate protein kinase activity (12), suggesting that dimerization may allosterically modulate kinase activity. Latent PKR exists predominantly as a monomer but does dimerize weakly, with Kd ~ 500 μM and dimerization at high protein concentrations in the absence of dsRNA is sufficient to activate PKR (16). Fusion of a heterologous dimerization domain with the PKR kinase domain enhances autophosphorylation (23, 24). These data support a model whereby dsRNA activates by binding multiple PKR monomers in close proximity, thereby enhancing dimerization via the kinase domain (14). Alternatively, RNA binding at the dsRBD may induce a conformation change in PKR monomers that enhances dimer stability and leads to subsequent dimerization (13). However, a defining feature of PKR activation by dsRNA is the “bell shaped” curve, where high RNA concentrations are inhibitory (25), which is not explained in the context of the latter model. In contrast, these observations are compatible with the former model, where inhibition would occur by dissociation of a PKR dimer residing on one dsRNA molecule into two monomers each bound to a separate RNA (26).
In order to differentiate among the alternative models for PKR activation it is necessary to define the nature of the interactions between domains and the conformational changes that accompany activation. The folding mechanisms and thermodynamic stabilities of multidomain proteins have been extensively studied (27–29). In general, the presence of large, densely packed domain interfaces results in stabilization of the neighboring domains and cooperative folding (28). Conversely, in multidomain proteins where the domains are connected by flexible linkers with no significant domain interfaces, the domains are thermodynamically and kinetically independent (30–32). Thus, one can gain insights into the interdomain contacts by comparing the relative stabilities of an isolated domain and the same domain expressed in the context of a multidomain protein. Here, we have compared the stabilities of full length PKR and its constituent domains using limited proteolysis and equilibrium chemical denaturation. PKR contains three tryptophans, all localized within the kinase domain, and we have used intrinsic tryptophan fluorescence as a probe to selectively detect unfolding of the kinase domain and CD spectroscopy to monitor the global unfolding reactions. In addition, we have used limited protease digestion to probe unstructured and accessible regions of PKR and to define conformational changes associated with cofactor binding and autophosphorylation. We find that the two dsRBM units fold independently, with dsRBM2 exhibiting greater stability. The kinase domain is slightly stabilized by the dsRBD, suggesting weak interaction between the domains. Protease digestion measurements indicate that autophosphorylation induces conformation changes that include structuring of the activation loop.
Materials and Methods
Reagents and Materials
All reagents used were reagent grade purchased from Fisher Scientific except as noted. Ultrapure urea was obtained from MP Biomedical, Trypsin was obtained from Sigma-Aldrich and chymotrypsin and papain were from Worthington Biochemical Corp. Solutions containing urea were purified by adding 5% w/v Biorad AG501-8X resin, swirling gently for 45 minutes and filtering.
Protein Expression
Expression and purification of the following PKR constructs has previous been described: Full-length PKR (16); dsRBD, encompassing both dsRBM1 and dsRBM2 (PKR 1–184) (33) and dsRBM1 (PKR 1–91) (34). The kinase domain (PKR 242–551) containing the K296R→R mutation was cloned into pET-11a and resulting plasmid was transformed into BL21 (DE3) E. coli expression cells (Novagen, Rosetta2 pLysS) Cells were grown in LB containing 50 μg/ml ampicillin or carbenicillin and 34 ug/ml chloramphenicol at 37°C until the OD at 600 nm = 0.7. The cells were cooled to 18°C and expression was induced by the addition of 1 mM isopropyl-1-thio-β-D-galacto-pyranoside. The cells were harvested 4–5 hours post-induction by centrifugation at 3,000 g for 10 minutes and were frozen at −80°C. The cell pellets were thawed at room temperature and resuspended in buffer A (20 mM HEPES, 50 mM NaCl, 0.1 mM EDTA, 10% glycerol (pH 7.5) 10 mM β-mercaptoethanol) containing protease inhibitor cocktail (Sigma-Aldrich) and were lysed by sonication. The lysate was centrifuged at 22,000 g for 15 minutes to pellet cell debris. The supernatant was loaded onto a Heparin Sepharose column (GE Healthcare) and kinase domain was eluted with a NaCl gradient. The peak fractions were pooled, concentrated to ~ 10 mg/ml and applied to a Superdex 75 column (GE Healthcare) that had been equilibrated in AU200 buffer (20 mM HEPES, 200 mM NaCl, 0.1 mM EDTA, 0.1 mM TCEP, pH 7.5). PKR autophosphorylation reactions were carried out at a protein concentration of 5 mg/ml in a buffer containing 50 mM Tris, 100 mM NaCl, 0.1 mM EDTA, 5 mM MgCl2, 10 mM BME, pH 8.0 and 5 mM ATP at room temperature for 4–5 hours. After incubation, the dimeric fraction was purified by gel filtration on a Superdex 200 column equilibrated in AU200 buffer.
Limited Proteolysis
PKR Proteolysis experiments were carried out using trypsin, chymotrypsin and papain. PKR (1 or 0.01 mg/ml) was incubated with variable amounts of protease for 30 minutes at 20°C. The reaction was quenched by adding SDS-PAGE sample buffer and heating to 90°C for 10 minutes. Proteolysis products were separated on 12% or 4–12% acrylamide bis-tris denaturing gels (Invitrogen) and visualized with Coomassie blue staining or silver staining. Cleavage sites were identified by transferring peptides onto a PVDF membrane for N-terminal sequencing at the Yale Keck facility.
Equilibrium chemical denaturation
Equilibrium urea denaturation studies were performed at 0.15–0.05 mg/ml protein in a buffer containing 10 mM HEPES, 200 mM NaCl, 0.1 mM EDTA, and 0.1 mM TCEP (pH 7.5). Samples were prepared at urea concentrations ranging from 0–8.5 M using a Hamilton Microlab 500 titrator and were equilibrated at 20°C for 3 hours. CD experiments were performed in an Applied Photophysics Pi Star 180 spectrapolarimeter at 20°C. Equilibrium titrations were recorded using a 5 mm pathlength cuvette monitoring the CD signal at 222 nm with entrance and exit slit widths of 3 nm. CD spectra were obtained with a 1 mm cuvette at 1 mg/ml protein concentration. Tryptophan fluorescence measurements were conducted with a Jobin Yvon Horiba FluoroMax-3 fluorimeter at 20°C. Tryptophan fluorescence changes were monitored using a 10 mm x 3 mm cell with an excitation wavelength of 295 nm (2 nm slit width) and an emission wavelength of 340 nm (4 nm slit width).
The CD and fluorescence data were globally fit to equilibrium transition models using the program SAVUKA (35, 36). The dependence of the observed signal on denaturant concentration is given by
| (1) |
where Yobs is the observed signal at a given denaturant concentration and YN and YU are the signals for the native and unfolded forms at the same denaturant concentration, and Fapp is the apparent fraction unfolded. Both YN and YU at a given denaturant concentration are obtained by making linear extrapolations from the baseline regions
| (2) |
where Y0N and Y0U represent the signals for native and unfolded states in the absence of urea, respectively, and sN and sU are the slopes that describe their denaturant dependence. For a simple two-state model, Fapp is given by
| (3) |
where [U] and [N] are the concentrations of the unfolded and native forms, respectively, and KNU is the equilibrium constant describing the N→U transition. For a 3-state transition involving an intermediate, I, Fapp is given by
| (4) |
where Z represents the spectroscopic properties of the intermediate and is given by Z = (YI−YN)/(YU−YN). The value of Z is zero when the intermediate resembles the native state and 1 when it resembles the unfolded state, as probed by a given optical system.
A linear dependence of the free energy of unfolding on urea concentration was assumed:
| (5) |
where ΔG°(D) is the standard free energy of unfolding in the presence of denaturant, ΔG°(H2O) is the standard free energy change in the absence of denaturant and m is a measure of the sensitivity of the apparent free energy change to denaturant concentration. In global analysis of the unfolding data, ΔG°(H2O) and m were treated as global parameters and Y0N, sN, Y0U, sU and Z were fit as local parameters.
Analytical ultracentrifugation
Sedimentation velocity analysis was conducted at 20°C and 55,000 RPM using interference optics with a Beckman-Coulter XL-I analytical ultracentrifuge. Double sector synthetic boundary cells equipped with sapphire windows were used to match the sample and reference menisci. Extinction coefficients, molecular masses, partial specific volumes and solvent densities were calculated using Sednterp (37). Initial analysis was performed using Sedfit (38) to obtain c(s) distributions and DCDT+ (39) to obtain g(s*) distributions. Multiple datasets were globally fit to hetero-association models using the programs Sedanal (40) and Sedphat (41).
Results
Limited proteolysis
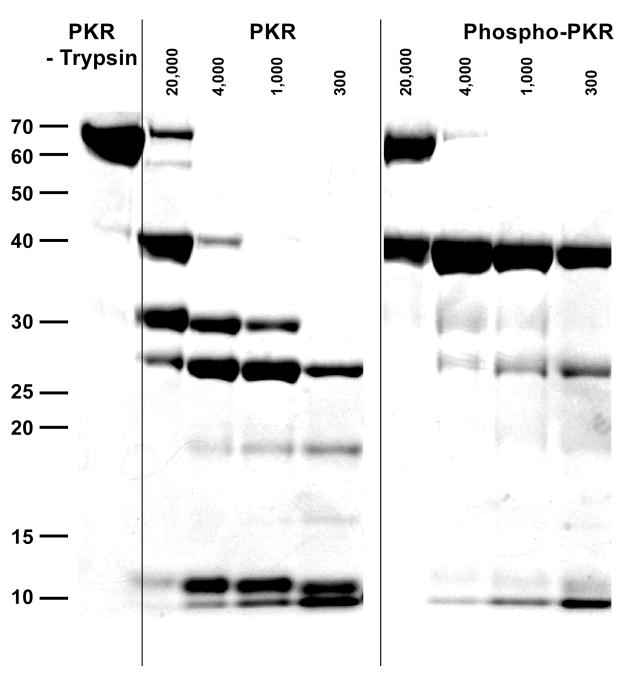
We have used limited proteolysis to define the accessible and unstructured regions of PKR and to monitor conformational changes associated with ligand binding and activation. Figure 2 shows SDS PAGE analysis of limited trypsin digests of PKR and phosphorylated PKR. At the lowest trypsin concentration, PKR is cleaved to generate two main peptides migrating as bands near 40 kDa and 30 kDa. N-terminal sequencing of these peptides reveals two nearby cleavage sites after R241 and R246 located in the linker region near the N-terminus of the kinase domain (Figure 1A, site A). This region is extremely susceptible to trypsin cleavage: addition of a 1:20,000 ratio of trypsin: PKR results in nearly complete cleavage in 30 minutes at 20°C. At higher [trypsin], the 40 kDa kinase domain peptide is cleaved and additional bands develop at 25 kDa and 11 kDa. Sequencing of these peptides reveals a cluster of nearby cleavage sites after R445, R447 and R453 located within the kinase domain activation loop (site B). At the highest trypsin concentrations, the 30 kDa fragment is cleaved to 18 kDa and 10 kDa products at K79 located between dsRBM1 and dsRBM2 (site C).
Figure 2.
Limited proteolysis of PKR. PKR was incubated at 1 mg/ml at several ratios of PKR: trypsin at 20 °C for 30 minutes. Reactions were quenched by the addition of SDS sample loading buffer and heating to 90°C for 10 minutes. The samples were run on a 4–12% acrylamide bis-tris gel under denaturing conditions and visualized with Coomassie blue staining. Lane 1 contains unphosphorylated PKR that has not been digested. Lanes 2–5 contain unphosphorylated PKR with increasing concentrations of trypsin. Lanes 6–9 contain phosphorylated PKR and increasing concentrations of trypsin.
We have investigated the effects of ligand binding and activation on the PKR cleavage reactions catalyzed by trypsin. The nonhydrolyzable ATP analogue AMPPNP binds to PKR with Kd ~ 100 μM (17). However, addition of 200 μM AMPPNP does not affect the trypsin proteolysis pattern. Similarly, binding of PKR to a 40 bp activating dsRNA or a 16 bp nonactivating RNA (34) also does not strongly perturb the proteolysis pattern. However, PKR autophosphorylation dramatically changes the proteolysis pattern (Figure 2). Although the initial cleavage at site A occurs, the resulting 40 kDa kinase domain fragment is stabilized against cleavage at site B within the activation loop. Interestingly, the 30 kDa dsRBD fragment is not detected in the phosphorylated samples, suggesting that the phosphorylation enhances proteolysis of this region of PKR by trypsin. Note that phosphorylation of PKR enhances dimerization such that Kd decreases from 450 to 0.95 μM (16). Thus, at the protein concentrations used in the proteolysis experiments, the unphosphorylated enzyme is predominantly monomeric whereas a significant fraction of the phosphoenzyme is dimeric. The 40 kDa kinase domain fragment from the phosphoenzyme is also resistant to proteolysis in measurements performed at 100-fold lower PKR concentration (0.01 mg/ml, 0.16 μM), indicating that the effect is not due to enhanced dimerization but is associated with phosphorylation.
Reversibility of PKR unfolding by urea
We carried out equilibrium urea denaturation measurements on full-length PKR and several domain constructs to define the thermodynamic stability and domain interactions. Thermodynamic interpretation of these experiments requires that the unfolding transitions are fully reversible. Control experiments were performed to demonstrate reversible unfolding of full-length PKR by comparing urea titrations starting from 0 M urea (fully folded) and 6.2 M urea (fully unfolded). These two samples were incubated at 20°C for 3 hours and then diluted to identical final urea and protein concentrations and equilibrated an additional 3 hours. The two titrations overlay closely, indicating that urea unfolds PKR reversibly without significant population of off pathway intermediates or aggregates (see supporting information). Because the entire multidomain protein unfolds reversibly it is reasonable to assume that the domain constructs also unfold reversibly.
dsRBD stability
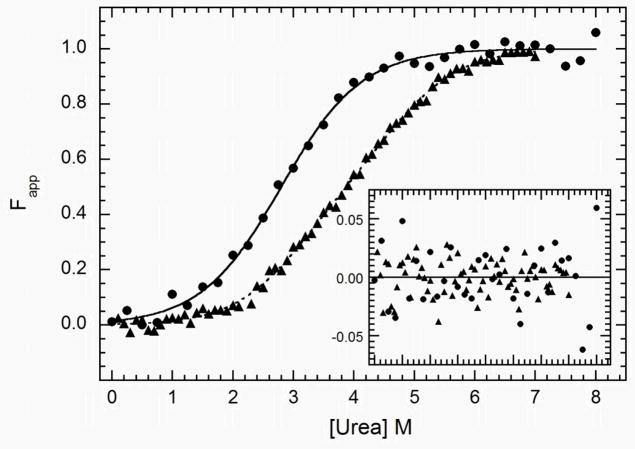
Equilibrium denaturation experiments were performed on several PKR domain constructs as well as the holoenzyme. Figure 3 shows titrations performed using two constructs encompassing dsRBM1 alone and a dsRBD construct encompassing both dsRNA binding motifs. Because the dsRBD does not contain any tryptophans, the unfolding reactions for these constructs were monitored with CD at 222 nm to detect α-helical content. When normalized and baseline-subtracted, the two datasets do not overlay, with the dsRBD unfolding reaction occurring over a broader range of urea concentrations and with a higher midpoint. As indicated by the random distribution of the residuals (Figure 3, inset), the dsRBM1 data fit well to a simple two state model with no intermediate and a stability of ΔG = 2.45 ± 0.48 kcal/mol and m = 0.88 ± 0.13 kcal mol−1 M−1. (Table 1). Attempts to fit the dsRBD data to a two-state model gave unreasonably low apparent stability and slopes (data not shown). However, a good fit to the dsRBD is obtained using a three state model incorporating an intermediate. Because the two motifs have the same fold, they are expected to contribute equally to the CD change upon unfolding and the Z-parameter was fixed at 0.5. The linker between the motifs is unstructured and does not contribute to the unfolding reaction. Within error, ΔGN-I for the dsRBD construct is equal to the stability of dsRBM1, but ΔGNI-U occurs with a significantly higher free energy change of 4.61 ± 0.71 kcal/mol. Thus, the N→I transition is assigned to unfolding of dsRBM1 and I→U with unfolding of dsRBM2, with the latter domain showing greater stability. A correlation has been established between m-values and the amount of surface area exposed upon chemical denaturation (42). Consistent with our assignments of the N→I and I→U transitions, the m-values for the two unfolding steps are equal and agree with that obtained for the dsRBM1 construct (Table 1). To confirm this interpretation, the dsRBM1 and dsRBD data were globally fit to the three-state model by fixing the Z values to 1.0 for dsRBM1 and 0.5 for dsRBD. The data fit well and the parameters derived from the global fit are in good agreement those obtained from separate fits to the two data sets.
Figure 3.
Analysis of the stability of dsRBM1 and dsRBD by equilibrium urea unfolding titrations. Samples were prepared at 0.1 mg/ml (dsRBM1) or 0.15 mg/ml (dsRBD) and incubated at variable urea concentrations at 20°C for 3 hours. CD data were collected at 222 nm in a 5 mm path length cuvette. The data were background-subtracted, normalized and fit with a two-state model (dsRBM1) or a three-state model (dsRBD) using the program SAVUKA. The points are the normalized data for dsRBM1 ( ) and dsRBD (
) and dsRBD ( ). and the lines are the respective two- and three-state fits. The inset shows the residuals. The best-fit parameters are in Table 1.
). and the lines are the respective two- and three-state fits. The inset shows the residuals. The best-fit parameters are in Table 1.
Table 1.
Equilibrium thermodynamic stability parameters.a
| Construct | ΔGN-I (kcal/mol) | mN-I (kcal mol−1 M−1) | ΔGI-U (kcal/mol) | mI-U (kcal mol−1 M−1) | Z (Fluorescence) | Z (CD) |
|---|---|---|---|---|---|---|
| dsRBM1 (CD) | 2.45 ± 0.48 | 0.88 ± 0.13 | ||||
| dsRBD (CD) | 3.01 ± 0.51 | 0.99 ± 0.17 | 4.61 ± 0.71 | 0.98 ± 0.17 | 0.5 (Fixed) | |
| dsRBM1 and dsRBD (CD)b | 2.95 ± 0.40 | 0.98 ± 0.13 | 4.56 ± 0.54 | 0.97 ± 0.12 | 1 (dsRBM1) and 0.5 (dsRBD) | |
| Kinase Domain (Fluorescence) | 5.17 ± 0.17 | 1.84 ± 0.06 | ||||
| Kinase Domain (CD+Fluorescence)c | 5.00 ± 0.25 | 1.76 ± 0.11 | 3.68 ± 1.25 | 1.01 ± 0.29 | 0.88 ± 0.07 | 0.34 ± 0.19 |
| PKR (Fluorescence) | 6.66 ± 0.53 | 2.19 ± 0.17 | ||||
| PKR (CD+Fluorescence)c | 6.10 ± 0.57 | 1.97 ± 0.19 | 2.74 ± 1.90 | 0.65 ± 0.49 | 1.00 ± 0.61 | 0.21 ± 0.44 |
| PhosphoPKR (Fluorescence) | 5.45 ± 0.28 | 1.84 ± 0.09 | ||||
| PhosphoPKR (CD+Fluorescence)c | 5.76 ± 0.35 | 1.91 ± 0.11 | 2.61 ± 0.44 | 0.53 ± 0.08 | 0.82 ± 0.09 | 0.26 ± 0.08 |
Parameters were obtained by fitting CD and fluorescence equilibrium urea denaturation curves using SAVUKA. PKR refers to the full length enzyme and PhosphoPKR refers to phosphorylated full length PKR. The dsRBM1 CD, kinase fluorescence, and PKR fluorescence data were fit to a two state model. All other data were fit to a three state model with one equilibrium intermediate. For details, see the text.
Global fit of the CD data for the dsRBM1 and dsRBD constructs.
Global fit of CD and fluorescence data.
Kinase domain stability
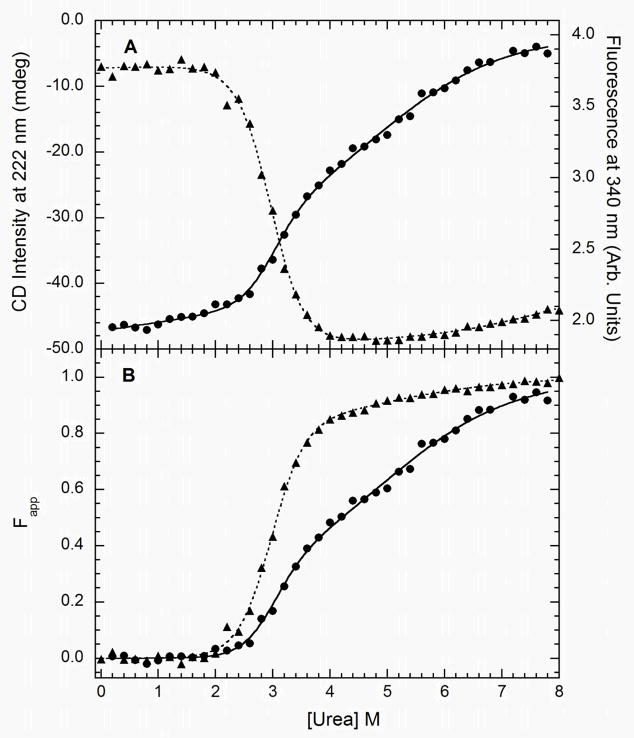
Based on the results of the limited proteolysis experiments above, our chemical denaturing measurements of the kinase domain utilized a construct encompassing PKR residues 242–551. The K296R mutation was introduced to prevent autophosphorylation during expression. Sedimentation velocity experiments were performed over a concentration range of 0.2 to 1.8 mg/ml to evaluate the association state of this construct. Analysis of these data using sedimentation coefficient distribution functions revealed a major peak at ~2.7 S that did not shift with loading concentration, indicating that the kinase does not undergo concentration-dependent self-association (data not shown). A small (3%) impurity is also observed near 4 S that is assigned to irreversible dimer. Sedimentation velocity data from a sample prepared at 0.6 mg/ml fit well to a discrete model with s20,w = 2.750 S and M= 35,653 Da. This molecular weight agrees well with predicted molar mass of 35,811 kDa, indicating that the kinase domain construct exists as a homogeneous monomer over the concentration range that was examined. The frictional ratio for the construct, f/f0 = 1.36, is in the range typically found for globular proteins.
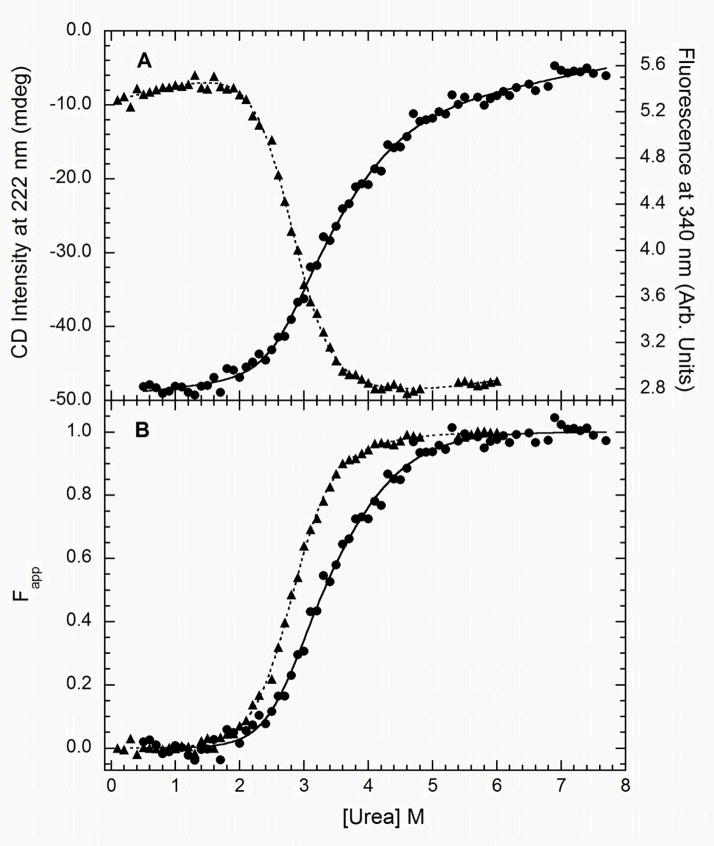
The kinase domain contains three tryptophans (Figure 1B) and urea-induced unfolding was monitored using a combination of CD and fluorescence spectroscopy. The tryptophan emission spectrum of PKR has a maximum at 347 nm which shifts to 355 nm and undergoes a 40% decrease in amplitude upon urea denaturation (see supplementary materials). The red-shift and quenching of the emission is associated with transfer of the tryptophan side chain from a hydrophobic environment to a solvent exposed state upon denaturation. Figure 4 shows the unnormalized (panel A) and normalized (panel B) CD and fluorescence titrations. The normalized CD and fluorescence data are not coincident, indicating the presence of at least one folding intermediate. The fluorescence change occurs at a lower urea concentration indicating that this signal predominantly monitors the N→ I transition. When analyzed alone, the fluorescence and CD data each fit well to a simple two-state model (Table 1) and the fit quality is not substantially improved with a three-state model incorporating an intermediate. As expected, global analysis of the fluorescence and CD data clearly requires a three-state model (Figure 4). The parameters for the N→ I transition correlate closely with those obtained from a two-state analysis of the fluorescence data alone and those associated with the N→ U step correspond to analysis of the CD data alone. The high value of Z = 0.88 ± 0.07 for fluorescence confirms that the tryptophan emission change occurs mostly in the N→ I step whereas Z = 0.34 ± 0.19 for CD indicates that the most substantial decrease in α-helical content happens at the I→ U transition. The first transition occurs with a higher free energy change and m-value than the second.
Figure 4.
Analysis of the stability of the kinase domain. Samples were prepared at 0.15 mg/ml and variable urea concentrations and incubated at 20°C for 3 hours. Unfolding was monitored using CD at 222 nm ( ) and tryptophan fluorescence emission with excitation at 295 and emission at 340 nm (
) and tryptophan fluorescence emission with excitation at 295 and emission at 340 nm ( ). A) Raw data. B) Background subtracted and normalized data. The lines are the global fit of the CD and fluorescence data to a three-state model obtained using SAVUKA.
). A) Raw data. B) Background subtracted and normalized data. The lines are the global fit of the CD and fluorescence data to a three-state model obtained using SAVUKA.
Stability of full length PKR
Equilibrium thermodynamic analysis of the stability of full length PKR holoenzyme was performed using CD and fluorescence titrations analogous to those performed above for the kinase domain. PKR exists in monomer-dimer equilibrium (16) and parallel experiments were performed at two protein concentrations of 0.15 mg/ml and 0.05 mg/ml to assess potential contributions of the association reaction to the unfolding thermodynamics. The CD titrations recorded at the two protein concentrations were coincident (data not shown) indicating that dimer dissociation does not contribute to the unfolding equilibria. Based on the measured Kd for PKR dimerization of ~ 500 μM, less that 1% dimer is populated at 0.15 mg/ml, so the absence of a concentration dependence is reasonable.
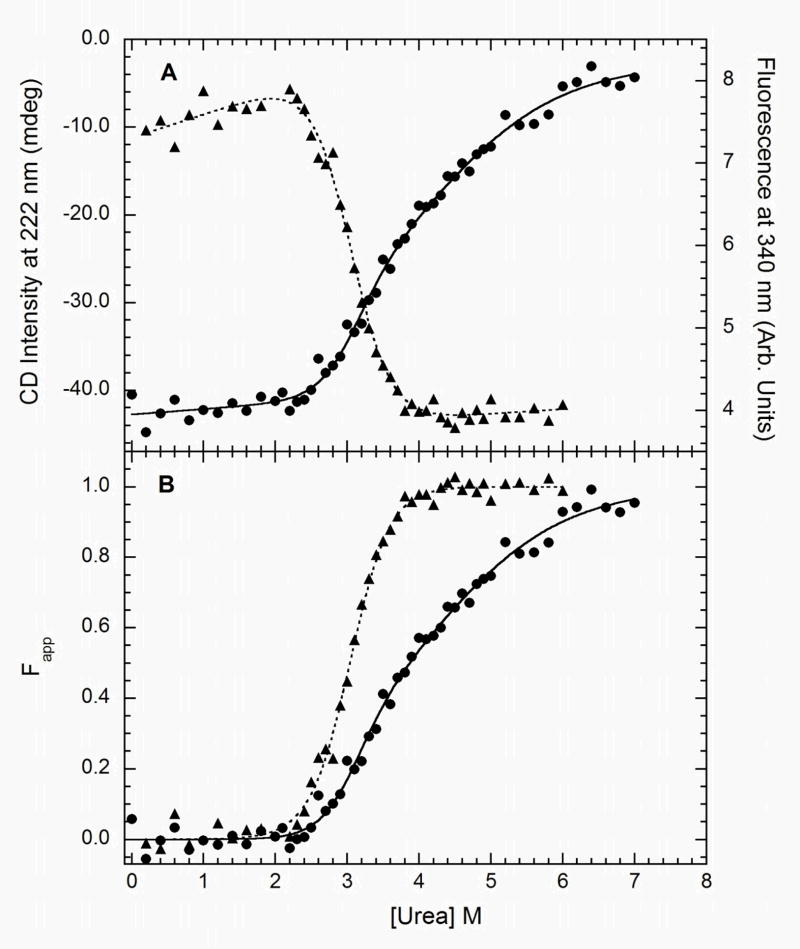
Figure 5 shows unnormalized and normalized urea denaturation curves for full length PKR. The data are qualitatively similar to those obtained with the isolated kinase domain. The fluorescence and CD data are clearly non-coincident, indicating the presence of at least one intermediate. The difference between the two signals is even more pronounced than was observed with the kinase domain. The CD curve is quite broad and a linear baseline in the unfolded region is not apparent at 7 M urea. Control CD titrations were carried out to higher urea concentrations (8.6 M) to verify the baseline slope in this region. Analysis of the unfolding thermodynamics of full length PKR is complicated by the presence of multiple overlapping transitions. Based on the analyses above, the kinase domain and dsRBD will each contribute two transitions. However, tryptophan fluorescence selectively reports on the N→ I transition of the kinase domain. Thus, we initially analyzed the fluorescence channel separately to compare stabilities of the kinase in the context of the isolated domain and full length PKR. As we observed for the kinase domain, the tryptophan fluorescence titration of full length enzyme fits well to a two state model. The free energy change of ΔG= 6.7 kcal/mol is about 1.5 kcal/mol larger than observed for the isolated kinase domain, suggesting that some stabilization of the kinase occurs in the context of the full length enzyme. The slightly higher m-value for PKR relative to the kinase domain construct indicates that the unfolding transition is coupled to exposure of a somewhat larger surface area.
Figure 5.
Analysis of PKR stability. Full length PKR was prepared at 0.15 mg/ml and variable urea concentrations and incubated at 20°C for 3 hours. Unfolding was monitored using CD at 222 nm ( ) and tryptophan fluorescence emission with excitation at 295 and emission at 340 nm (
) and tryptophan fluorescence emission with excitation at 295 and emission at 340 nm ( ). A) Raw data. B) Background subtracted and normalized data. The lines are the global fit of the CD and fluorescence data to a three-state model obtained using SAVUKA.
). A) Raw data. B) Background subtracted and normalized data. The lines are the global fit of the CD and fluorescence data to a three-state model obtained using SAVUKA.
The resolution of the urea titrations for PKR is not sufficient to reliably extract parameters for all of the transitions that are expected to occur upon unfolding of this multidomain protein. The fluorescence and CD titrations were globally analyzed using both three-state and four-state models. However, fits to the three state model gave random residuals and there was no improvement in fit quality for the four-state model. Thus, only the three-state fits are presented in table 1. The free energy change for the N→ I transition in this fit is slightly lower than obtained from independent analysis of the fluorescence data. The poorer agreement between the single channel and global analysis relative to what is obtained upon analysis of the kinase domain data likely reflects the contribution of the dsRBD unfolding transitions to the data for the full length enzyme. Thus, the three-state global analysis of full length PKR unfolding should be viewed as a phenomenological description of the data rather than a thermodynamic dissection of the complete unfolding pathway. For example, the low m-value for the I → U reaction of 0.64 kcal mol−1 M−1 is likely an artifact due to broadening by overlapping transitions. The value of Z=1.0 for the fluorescence channel does confirm that the N→ I unfolding reaction of the kinase occurs early in the titration.
Chemical denaturation of the phosphorylated form of PKR was also characterized to determine whether activation induces a global change in stability in addition to the conformation change in the activation loop detected by limited proteolysis. As with latent PKR, we detect no concentration dependence for the unfolding reaction of the phosphoenzyme (data not shown). Dimerization of PKR is greatly enhanced by phosphorylation, and based on the measured Kd of 0.95 μM (16) the enzyme is 65% dimeric at 0.15 mg/ml. Thus, the absence of any protein concentration dependence to the unfolding reactions monitored by CD and fluorescence changes is unexpected. However, given the complexity of this multidomain unfolding it is possible that a concentration-dependent step is masked by other overlapping transitions.
Figure 6 shows fluorescence and CD titrations for the phosphoenzyme. The CD signal change is very broad, suggesting the presence of multiple overlapping transitions. The fluorescence change fits well to simple two-state model with a free energy of 5.45 ± 0.28 kcal/mol (Table I). Comparing this value to those obtained from analysis of the florescence titrations of the other constructs indicates that phosphorylation destabilized the kinase domain in full length PKR by about 1 kcal/mol such that it is only slightly more stable than the isolated kinase. As in the case of the latent enzyme, even with global analysis of fluorescence and CD data it is not possible to resolve the each of the intermediates that are expected to be populated upon unfolding of phosphorylated PKR.
Figure 6.
Analysis of the stability of phosphorylated PKR stability. Full length PKR was prepared at 0.15 mg/ml and variable urea concentrations and incubated at 20°C for 3 hours. Unfolding was monitored using CD at 222 nm ( ) and tryptophan fluorescence emission with excitation at 295 and emission at 340 nm (
) and tryptophan fluorescence emission with excitation at 295 and emission at 340 nm ( ). A) Raw data. B) Background subtracted and normalized data. The lines are the global fit of the CD and fluorescence data to a three-state model obtained using SAVUKA.
). A) Raw data. B) Background subtracted and normalized data. The lines are the global fit of the CD and fluorescence data to a three-state model obtained using SAVUKA.
Sedimentation velocity analysis of the interaction of kinase and dsRBD constructs
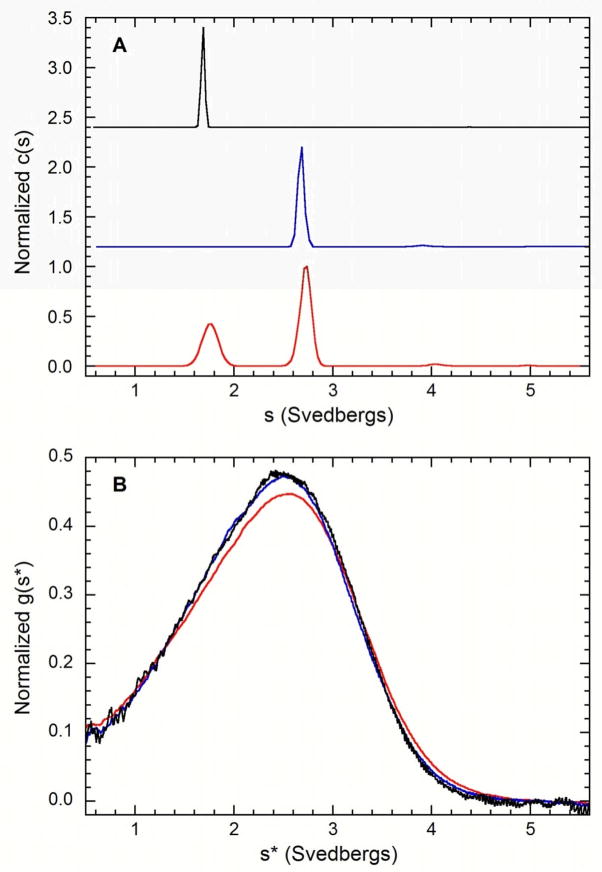
Previous NMR chemical shift perturbation experiments indicate that the PKR kinase domain and dsRBD construct interact (10, 15). We have quantitatively examined this interaction using sedimentation velocity experiments. Data were initially analyzed using continuous sedimentation coefficient [c(s)] distributions. This method removes diffusional broadening and is useful to define the species present in an unknown sample. Figure 7A shows c(s) distributions for samples containing kinase domain, dsRBD and a 1:1 (molar) mixture of these constructs. As indicated above, the kinase domain sediments as an s = 2.7 S, monomeric species. The dsRBD is also monomeric (34), with s ~ 1.7 S. The same features associated with the kinase domain and dsRBD monomeric species are observed in the c(s) distribution of the mixture but no additional higher S species are detected, indicating that a stable kinase-dsRBD complex is not formed at appreciable concentrations under these conditions.
Figure 7.
Sedimentation velocity analysis of the interaction of dsRBD and kinase domains. A) Continuous sedimentation coefficient distribution analysis of dsRBD (black), kinase domain (blue) and 1:1 (molar) mixture of dsRBD and kinase domain (red). The distributions are normalized by peak height and offset for clarity. The small feature near 4 S in the samples containing kinase domain is assigned as an irreversible kinase dimer contaminant. Conditions: sample concentrations, 0.5 mg/ml (dsRBD), 0.5 mg/ml (kinase domain), 1 mg/ml (mixture); rotor speed, 55,000 RPM; temperature, 20°C; interference optics. B) g(s*) analysis of 1:1 mixtures of dsRBD: kinase domain at concentrations of 3.2 mg/ml (red), 1.1 mg/ml (blue) and 0.36 mg/ml (black). The distributions are normalized by concentration. Conditions: rotor speed, 55,000 RPM; temperature, 20°C; interference optics.
We have tested for weak complex formation by comparing the sedimentation velocity data obtained from 1:1 kinase-dsRBD mixtures prepared at several dilutions. The data were analyzed using normalized g(s*) distributions to facilitate comparison of data obtained at different concentrations. Figure 7B shows the overlay of distributions obtained from a mixture containing 50 μM kinase domain and dsRBD (3.2 mg/ml total protein) along with 1:3 and 1:9 dilutions of this mixture. Although the resolution of the kinase domain and dsRBD is reduced by diffusional broadening in this analysis method, there is a clear shift of the distributions to the left in the 1:3 dilution which persists in the 1:9 dilution. The weight-average sedimentation coefficients obtained by integration of these distributions decreases from 2.335 ± 0.001 S at the highest concentration to 2.307 ± 0.002 S in the 1:3 diluted samples. This shift to lower sedimentation coefficient upon dilution is consistent with a weakly associating system in rapid exchange on the timescale of the sedimentation experiment. Note that potential complications from hydrodynamic and thermodynamic nonideality preclude sedimentation experiments at higher protein concentrations.
Finally, whole-boundary global fitting methods were used to estimate the Kd of the interaction using four datasets obtained at multiple ratios of kinase: dsRBD and loading concentrations. A 1:1 kinase:dsRBD stoichiometry for the complex was assumed. The sedimentation coefficients for the monomers were fixed at the experimentally determined values. Because very little of the complex is populated, even at the highest loading concentration, it was not possible to treat the sedimentation coefficient of the complex as an adjustable parameter. Although the frictional properties of the complex are not known, it is reasonable to assume that s will lie between the values obtained for the kinase domain alone (2.7 S) and full-length PKR (3.5 S). Constraining s to lie between 3.0 and 3.5 S gave a best fit values of Kd between 163–307 μM with RMSD = 0.021 fringes (data not shown). The data fit significantly less well to a noninteracting mixture model with RMSD = 0.035 fringes, indicating that the interacting model is a better description of the data and the kinase domain does bind to dsRBD, albeit with low affinity.
Discussion
Analysis of the thermodynamic stability of PKR and its constituent domains by equilibrium chemical denaturation and limited proteolysis experiments indicates limited interaction between domains. Consistent with previous NMR evidence that dsRBM1 and dsRBM2 do not interact (9, 10), we find that their folding energetics are noncooperative and the linker connecting them is accessible to cleavage by trypsin. Although the overall structures of the two domains are quite similar, with a backbone RMSD of 2 Å (9), dsRBM2 is about 1.6 kcal/mole more stable than dsRBM1. dsRBM1 does exhibit significantly more backbone motion on the millisecond timescale than dsRBM2 (10) and this conformational flexibility may be correlated with reduced thermodynamic stability.
Global analysis of PKR kinase domain unfolding monitored by CD and tryptophan fluorescence reveals the presence of an equilibrium intermediate. The fluorescence change occurs almost exclusively at the N → I transition. In contrast, the Z-value of 0.34 for CD implies a substantial loss of helical content is associated with both transitions. The kinase domain contains three tryptophans: W327 lies in the β-sheet in the N lobe, W377 is in helix αD on the C-lobe and W539 is near the bottom of the C-lobe on helix αJ (Figure 1B). Calculations of the side chain solvent accessible surface areas (43) indicate that each of the three tryptophans are substantially buried, with only 7%, 17% and 5% solvent exposure, respectively. Thus, it is not possible to assign the observed fluorescence change to one of the tryptophans based on their changes in solvent exposure upon denaturation. In two receptor tyrosine kinase domains, a single conserved tryptophan located in the catalytic cleft dominates the fluorescence emission spectrum (44, 45). However, this tryptophan is not conserved in Ser/Thr protein kinases such as PKR.
Because PKR tryptophan fluorescence changes reports on an unfolding transition localized in the kinase domain, this signal can be used to assess the effects of domain interactions on the stability of the kinase domain. Using this approach, we find that the kinase domain is slightly stabilized by ~1.5 kcal/mole in the context of the holoenzyme. In multidomain proteins, the degree of domain stabilization correlates with the size of the interface (28) and domains that are only connected by flexible linkers fold independently (30–32). Thus, the small stabilization we observe of the kinase domain of PKR implies that the catalytic domain does not interact extensively with other regions of the enzyme. The slight stabilization is reversed upon phosphorylation. This reduction in stability may be associated with reduced interaction of the kinase domain with other regions in the phosphorylated enzyme or may reflect a loss of intrinsic stability of the kinase domain. Consistent with the latter explanation, phosphorylation of the isolated insulin receptor kinase decreases its stability by about 2 kcal/mole (44).
Sedimentation velocity experiments indicate that the dsRBD binds weakly to the kinase. No discrete peak associated with a dsRBD-kinase complex is observed in the c(s) distributions of mixtures of these constructs, but the shift of the g(s*) distributions to lower s upon dilution indicates weak, reversible interaction. Based on global analysis of the sedimentation velocity traces we estimate Kd ~250 μM. Previously, a Kd of 0.5 μM was determined by surface plasmon resonance (10) and Kd < 100 μM was reported based on NMR titrations (15). The origin of these discrepancies is not clear. The surface plasmon resonance measurements were performed using a maltose binding protein PKR kinase fusion construct and experimental conditions were not reported. The Kd < 100 μM from NMR was estimated as the lowest dsRBD concentration that affects the NMR spectrum of the kinase at 50 μM. This estimate is dependent on sensitivity and the magnitude of the chemical shifts.
The 80 amino acid linker region lying between the dsRBD and kinase domain is reported to be unstructured (13) and thus may serve as a flexible tether to enhance this interaction in the intact enzyme. We can estimate the intramolecular binding constant using the expression
| (6) |
where Ki is the intramolecular binding constant, K is the measured intermolecular binding constant (~4×103 M−1) and p(d) is the probability density for the peptide linker to have an end-to-end distance of d (46). The parameter p(d) can be treated as an effective concentration. Although the structure for the bound state is not known, NMR chemical shift perturbation measurements indicate that dsRBM2 binds to a region in the C-lobe of the kinase (15), and modeling a range of potential geometries indicates that d lies between 30–60 Å. Evaluating p(d) using a worm-like chain model for the linker (47) gives a range of effective concentrations of ~ 0.3 – 3 mM and thus the equilibrium constant Ki ~1– 10. These calculations suggest that PKR exists in an equilibrium between closed and open states and are consistent with our previous AFM results (17) where we observed both closed and open conformations and our hydrodynamic (16) data indicating an extended shape. However, the fact that Ki lies close to 1 argues against the autoinhibition model, where latent PKR is locked in a stable, closed conformation and dsRBD interacts with the kinase domain to block substrate binding.
Limited proteolysis results support the previously defined domain structure of PKR and provide new insights into the activation mechanism. As expected, some cleavages occur at domain boundaries. In particular, the N -terminus of the kinase is extremely susceptible to trypsin hydrolysis at R241 and R246, and this region is also readily cleaved by papain and chymotrypsin (E. Anderson, unpublished observations). The sensitivity of this region to proteolysis may be physiologically significant. PKR is specifically cleaved by caspases at D251 upon stimulation of apoptosis, resulting in enhanced phosphorylation of eIF2α and inhibition of protein synthesis (48). We also observe that the linker lying between dsRBM1 and dsRBM2 can be cleaved by trypsin at K79, consistent with previous evidence that this region is flexible and unstructured (9, 10). Interestingly, these interdomain cleavage reactions are not appreciably affected upon binding ATP analogues or dsRNA activators. These results suggest that binding of nucleotides or activators does not induce large scale domain rearrangements that would be expected to modulate accessibility to trypsin hydrolysis.
A cluster of proteolysis sites at R445, R447 and R453 lie within the activation loop of the PKR kinase domain. The activity of many kinases is modulated by phosphorylation in the activation loop, leading to an ordered conformation associated with a catalytically competent state (12, 49). Multiple autophosphorylation sites have been identified in PKR, including T446 (50, 51) and possibly T451 (52). The PKR activation loop adopts the canonical, ordered conformation in the crystal structure of PKR kinase domain phosphorylated at T446 (11). Although cleavage of the activation loop is not affected by substrate or activator binding it is strongly inhibited by autophosphorylation. We propose that phosphorylation at T446 and/or T451 induces an ordered active loop conformation that is resistant to trypsin digestion. The affinity and kinetics of nucleotide binding to PKR are not affected by phosphorylation state (17), indicating that the conformational change in the activation loop does not alter active site accessibility but may instead affect the kinetics of phosphoryl transfer.
Supporting Information
Figure S1: Limited proteolysis of PKR at low concentration.
Figures S2: Reversibility of urea-induced denaturation of PKR
Figure S3: Effect of urea-induced unfolding on the PKR tryptophan emission spectrum.
This material is available free of charge via the internet at http://pubs.acs.org.
Acknowledgments
We thank Carolyn Teschke for use of the titrator and CD instrumentation and for helpful suggestions and Osman Bilsel for the SAVUKA software and for assistance in the analysis of equilibrium unfolding data.
Abbreviations
- CD
circular dichroism
- dsRBD
dsRNA binding domain
- dsRBM
dsRNA binding motif
- dsRBM1
dsRNA binding motif 1 consisting of PKR residues 6–79
- dsRBM2
dsRNA binding motif 2 consisting of PKR residues 96–169
- dsRNA
double-stranded RNA
- eIF2α
eukaryotic initiation factor 2α
- TCEP
Tris[2-carboxyethyl] phosphine
Footnotes
This work was supported by grant AI-53615 from NIH (J.L.C.)
References
- 1.Clemens MJ, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman RJ. The double stranded RNA-activated protein kinase PKR. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. pp. 503–528. [Google Scholar]
- 3.Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- 4.Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Williams BR. Signal integration via PKR. Sci STKE. 2001:RE2. doi: 10.1126/stke.2001.89.re2. [DOI] [PubMed] [Google Scholar]
- 6.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 8.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded RNA binding motif: Interference and much more. Nature Rev Mol Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 9.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nanduri S, Rahman F, Williams BRG, Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 12.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 13.McKenna SA, Lindhout DA, Kim I, Liu CW, Gelev VM, Wagner G, Puglisi JD. Molecular framework for the activation of RNA-dependent protein kinase. J Biol Chem. 2007;282:11474–1186. doi: 10.1074/jbc.M700301200. [DOI] [PubMed] [Google Scholar]
- 14.Cole JL. Activation of PKR: an open and shut case? Trends Biochem Sci. 2007;32:57–62. doi: 10.1016/j.tibs.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelev V, Aktas H, Marintchev A, Ito T, Frueh D, Hemond M, Rovnyak D, Debus M, Hyberts S, Usheva A, Halperin J, Wagner G. Mapping of the auto-inhibitory interactions of protein kinase R by nuclear magnetic resonance. J Mol Biol. 2006;364:352–363. doi: 10.1016/j.jmb.2006.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemaire PA, Lary J, Cole JL. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J Mol Biol. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 17.Lemaire PA, Tessmer I, Craig R, Erie DA, Cole JL. Unactivated PKR exists in an open conformation capable of binding nucleotides. Biochemistry. 2006;45:9074–9084. doi: 10.1021/bi060567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carpick BW, Graziano V, Schneider D, Maitra RK, Lee X, Williams BRG. Characterization of the solution complex between the interferon-induced double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J Biol Chem. 1997;272:9510–9516. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 19.Gabel F, Wang D, Madern D, Sadler A, Dayie K, Daryoush MZ, Schwahn D, Zaccai G, Lee X, Williams BR. Dynamic flexibility of double-stranded RNA activated PKR in solution. J Mol Biol. 2006;359:610–623. doi: 10.1016/j.jmb.2006.03.049. [DOI] [PubMed] [Google Scholar]
- 20.Langland JO, Jacobs BL. Cytosolic double-stranded RNA-dependent protein kinase is likely a dimer of partially phosphorylated Mr=66,000 subunits. J Biol Chem. 1992;267:10729–10736. [PubMed] [Google Scholar]
- 21.Patel RC, Stanton P, McMillan NM, Williams BR, Sen GC. The interferon-inducible double-stranded RNA-activated protein kinase self-associates in vitro and in vivo. Proc Natl Acad Sci USA. 1995;92:8283–8287. doi: 10.1073/pnas.92.18.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson HD, Mathews MB. The regulation of the protein kinase PKR by RNA. Biochimie. 1996;78:909–914. doi: 10.1016/s0300-9084(97)86712-0. [DOI] [PubMed] [Google Scholar]
- 23.Ung TL, Cao C, Lu J, Ozato K, Dever TE. Heterologous dimerization domains functionally substitute for the double-stranded RNA binding domains of the kinase PKR. EMBO J. 2001;20:3728–3737. doi: 10.1093/emboj/20.14.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vattem KM, Staschke KA, Wek RC. Mechanism of activation of the double-stranded-RNA-dependent protein kinase, PKR: role of dimerization and cellular localization in the stimulation of PKR phosphorylation of eukaryotic initiation factor-2 (eIF2) Eur J Biochem. 2001;268:3674–3684. doi: 10.1046/j.1432-1327.2001.02273.x. [DOI] [PubMed] [Google Scholar]
- 25.Hunter T, Hunt T, Jackson RJ, Robertson HD. The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J Biol Chem. 1975;250:409–417. [PubMed] [Google Scholar]
- 26.Kostura M, Mathews MB. Purification and activation of the double-stranded RNA-dependent eIF-2 kinase DAI. Mol Cell Biol. 1989;9:1576–1586. doi: 10.1128/mcb.9.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garel JR. Folding of large proteins: multidomain and multisubunit proteins. In: Creighton TE, editor. Protein Folding. W. H. Freeman and Company; New York: 1992. pp. 405–454. [Google Scholar]
- 28.Han JH, Batey S, Nickson AA, Teichmann SA, Clarke J. The folding and evolution of multidomain proteins. Nat Rev Mol Cell Biol. 2007;8:319–330. doi: 10.1038/nrm2144. [DOI] [PubMed] [Google Scholar]
- 29.Jaenicke R, Lilie H. Folding and association of oligomeric and multimeric proteins. Adv Protein Chem. 2000;53:329–401. doi: 10.1016/s0065-3233(00)53007-1. [DOI] [PubMed] [Google Scholar]
- 30.Scott KA, Steward A, Fowler SB, Clarke J. Titin; a multidomain protein that behaves as the sum of its parts. J Mol Biol. 2002;315:819–829. doi: 10.1006/jmbi.2001.5260. [DOI] [PubMed] [Google Scholar]
- 31.Steward A, Adhya S, Clarke J. Sequence conservation in Ig-like domains: the role of highly conserved proline residues in the fibronectin type III superfamily. J Mol Biol. 2002;318:935–940. doi: 10.1016/S0022-2836(02)00184-5. [DOI] [PubMed] [Google Scholar]
- 32.Zarnt T, Tradler T, Stoller G, Scholz C, Schmid FX, Fischer G. Modular structure of the trigger factor required for high activity in protein folding. J Mol Biol. 1997;271:827–837. doi: 10.1006/jmbi.1997.1206. [DOI] [PubMed] [Google Scholar]
- 33.Ucci JW, Cole JL. Global Analysis of Nonspecific Protein-Nucleic Interactions by Sedimentation Equilibrium. Biophys Chem. 2004;108:127–140. doi: 10.1016/j.bpc.2003.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ucci JW, Kobayashi Y, Choi G, Alexandrescu AT, Cole JL. Mechanism of interaction of the double-stranded RNA (dsRNA) binding domain of protein kinase R with short dsRNA sequences. Biochemistry. 2007;46:55–65. doi: 10.1021/bi061531o. [DOI] [PubMed] [Google Scholar]
- 35.Bilsel O, Zitzewitz JA, Bowers KE, Matthews CR. Folding mechanism of the alpha-subunit of tryptophan synthase, an alpha/beta barrel protein: global analysis highlights the interconversion of multiple native, intermediate, and unfolded forms through parallel channels. Biochemistry. 1999;38:1018–1029. doi: 10.1021/bi982365q. [DOI] [PubMed] [Google Scholar]
- 36.Gualfetti PJ, Bilsel O, Matthews CR. The progressive development of structure and stability during the equilibrium folding of the alpha subunit of tryptophan synthase from Escherichia coli. Protein Sci. 1999;8:1623–1635. doi: 10.1110/ps.8.8.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, Horton J, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge: 1992. pp. 90–125. [Google Scholar]
- 38.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 40.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 41.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 42.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comp Chem. 1998;19:319–333. [Google Scholar]
- 44.Bishop SM, Ross JB, Kohanski RA. Autophosphorylation dependent destabilization of the insulin receptor kinase domain: tryptophan-1175 reports changes in the catalytic cleft. Biochemistry. 1999;38:3079–3089. doi: 10.1021/bi982546s. [DOI] [PubMed] [Google Scholar]
- 45.Lee S, Lin X, McMurray J, Sun G. Contribution of an active site cation-pi interaction to the spectroscopic properties and catalytic function of protein tyrosine kinase Csk. Biochemistry. 2002;41:12107–12114. doi: 10.1021/bi026439g. [DOI] [PubMed] [Google Scholar]
- 46.Zhou HX. Polymer models of protein stability, folding, and interactions. Biochemistry. 2004;43:2141–2154. doi: 10.1021/bi036269n. [DOI] [PubMed] [Google Scholar]
- 47.Zhou HX. Loops in proteins can be modeled as worm-like chains. J Phys Chem B. 2001;105:6763–6766. [Google Scholar]
- 48.Saelens X, Kalai M, Vandenabeele P. Translation inhibition in apoptosis: caspase-dependent PKR activation and eIF2-alpha phosphorylation. J Biol Chem. 2001;276:41620–41628. doi: 10.1074/jbc.M103674200. [DOI] [PubMed] [Google Scholar]
- 49.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 50.Romano PR, Garcia-Barrio MT, Zhang X, Wang Q, Taylor DR, Zhang F, Herring C, Mathews MB, Qin J, Hinnebusch AG. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol Cell Biol. 1998;18:2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Herring CJ, Romano PR, Szczepanowska J, Brzeska H, Hinnebusch AG, Qin J. Identification of phosphorylation sites in proteins separated by polyacrylamide gel electrophoresis. Anal Chem. 1998;70:2050–2059. doi: 10.1021/ac971207m. [DOI] [PubMed] [Google Scholar]
- 52.Zhang F, Romano PR, Nagamura-Inoue T, Tian B, Dever TE, Mathews MB, Ozato K, Hinnebusch AG. Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J Biol Chem. 2001;276:24946–24958. doi: 10.1074/jbc.M102108200. [DOI] [PubMed] [Google Scholar]