Abstract
Background and Aims
Mycorrhizal associations are essential to the plant kingdom. The largest flowering plant family, the Orchidaceae, relies on mycorrhizal fungi for germination, growth and survival. Evidence suggests varying degrees of fungal-host specificity based on a single fungal isolate from a single plant. This paper shows for the first time the diversity of endophytes colonizing in a single plant over consecutive years and the functional significance of this diversity.
Methods
Stem-collars of Caladenia formosa were collected in different seasons and years. Mycorrhizal fungi isolated were tested for their efficacy to induce leafing and genetically determined using ITS-RFLP and sequencing.
Results
Multiple mycorrhizal fungi were repeatedly isolated from a single collar that displayed varying effectiveness in germination percentages and adult leaf length. Additional factors contributed to the isolation of effective mycorrhizal fungi; fungal collection season, year of collection and individual isolates. Surface sterilization only improved the number of isolated mycorrhizal fungi. Dual inoculation did not increase germination. All 59 mycorrhizal fungi effective in germinating seed belonged to one clearly defined ITS (internal transcribed spacer) clade and clustered close to Sebacina vermifera (79–89 % homology). Isolates resulting in the greatest germination were not necessarily those resulting in the greatest survival and growth 1 year after germination.
Conclusion
Single orchid plants contained multiple mycorrhizal fungal strains of one species that had diverse functional differences. These results suggest that our current knowledge of fungal–host specificity may be incomplete due to experimental and analytical limitations. It also suggests that the long-term effectiveness of a mycorrhizal fungus or fungi could only be found by germination and longer-term growth tests rather than genetically.
Key words: Mycorrhizal fungi, genetic diversity, effectiveness, germination, isolation, ITS, seasonal influences, Caladenia formosa, Orchidaceae
INTRODUCTION
With over 92 % of land plants requiring mycorrhizal associations for carbon (Gebauer and Meyer, 2003), nitrogen (Cameron et al., 2006) and phosphorus (Cameron et al., 2007), symbiotic relationships are essential for plant survival (Wang and Qiu, 2006). The largest flowering plant family, the Orchidaceae, relies heavily on mycorrhizal fungi; with some achlorophyllous and terrestrial temperate orchids totally dependent on fungi (Julou et al., 2005; Leake, 2005; Dearnaley, 2007; Waterman and Bidartondo, 2008). Caladenia spp. are terrestrial temperate orchids confined to Australasia that are unusual in that they generally lack roots and the symbiont(s) colonize(s) the base of the single leaf (the stem-collar as defined by Ramsay et al., 1986). For endangered orchids, including C. formosa, the effectiveness of the mycorrhizal symbiosis is vital for their development and survival. It is essential to obtain an effective mycorrhizal fungus that cannot only germinate seeds but also maintain robust growth of the orchid for long-term survival ex situ.
Symbiotic relationships are complex. Different orchid developmental stages (germination and growth) may require different fungi (Rasmussen, 1995; Sharma et al., 2003; Huynh et al., 2004). It has been suggested that orchids associate with multiple symbionts even within a single peloton (Kristiansen et al., 2001a; Raleigh et al., 2001; McKendrick et al., 2002; Taylor et al., 2003), a single root (Taylor et al., 2003), a single plant (Bougoure et al., 2005) or a single population (McKendrick et al., 2002). Different microorganisms, including symbiotic and non-symbiotic fungi and bacteria, may be isolated from orchids (Wilkinson et al., 1989; Currah et al., 1990), but it is not clear if they are truly endophytic. Some authors surface-sterilize and some do not, and the relative abundance of the fungi isolated may vary with orchid developmental phase and season. The most common mycorrhizal fungi in orchids are in the Rhizoctonia alliance (Pope and Carter, 2001), and so all slow-growing and hyaline fungal isolates are considered as potential symbionts, whereas dark or fast-growing fungi are typically pathogenic (Warcup, 1981). Huynh et al. (2004) suggested that the appearance of orchid endophytes varied during a season, but they may also vary in effectiveness and so knowing when best to collect collars may be of vital importance in recovery plans.
The fungus species reported as being symbiotic with Caladenia species is Sebacina vermifera Oberw. (synonymous with Serendipita vermifera Oberw. P. Roberts) (Warcup, 1971). Warcup isolated almost exclusively this fungus from 20 Caladenia species and identified it on the basis of its sexual sporulation (Warcup and Talbot, 1967). Mycorrhizal fungi with similar morphology have been isolated since then from Caladenia species, but their identification has been problematic because sexual (telomorphic) stages has yet to be induced or observed (Clements, 1981; Ramsay et al., 1986). Until recently, they have only been numbered and named tentatively on the basis of asexual characteristics. More recently, however, molecular techniques have been used to show that some at least are closely allied to S. vermifera (Bougoure et al., 2005), using as a reference the ITS nrDNA region of one of Warcup's cultures (Warcup, 1981) of S. vermifera from C. dilatata (http://www.ncbi.nlm.nih.gov/; AF202728). This has the potential for use as a reference to allow other researchers to compare similar fungi from closely related Caladenia species or to elucidate their identity.
The aims were to elucidate the diversity of endophytes associating with C. formosa over consecutive years and the functional significance of this diversity. This study investigated (a) whether one or more symbionts(s) colonized C. formosa within each peloton, plant, season and year, (b) the relative proportions of microorganisms and the effectiveness of mycorrhizal isolates from C. formosa, (c) if surface-sterilization made a difference to success in isolating mycorrhizal fungi free from contamination, (d) if isolates that were most effective in germinating seeds were also most effective in longer-term orchid growth, and (e) if effective and ineffective isolates could be differentiated on a molecular basis.
MATERIALS AND METHODS
Plant collection
Caladenia formosa perennates as a dormant underground tuber over summer, emerges with a single leaf during late autumn, flowers and sets seed in spring before dying down again over summer. Eight plants were collected in 2000 and four in 2001 from the sole site, in eucalypt woodland on sandy soils in south-western Victoria, with generally moist winters and dry summers (Huynh et al., 2004). Collections were extremely limited since this species is endangered. In 2000, one plant was collected at each of the six main developmental orchid phases: leafing, budding, flowering, fruiting, senescing and dormancy (except that three plants were collected at fruiting), but, in 2001, plants were collected only at the first four phases, due to the lack of success in isolating effective mycorrhizal fungi after fruiting in 2000.
Plants were washed in running tap water and the stem-collar (swollen area at the leaf base) was excised. For surface-sterilized treatments, collars were immersed in 1 % NaOCl for 3 min before being washed in five changes of sterile water; for non-surface-sterilized stem-collars, the NaOCl treatment was omitted. Only single pelotons that were opaque, circular-oval and submersible were cultured, as described previously (Huynh et al., 2004). In 2000, 182 pelotons (26 from each plant at each phase) were selected randomly and transferred to individual plates of solid Pa5 medium (Ramsay et al., 1986) using a separate sterile Pasteur pipette for each peloton (Huynh et al., 2004). In 2001, 130 pelotons were so cultured: 26 pelotons from each of leafing, budding, flowering (surface-sterilized and not surface-sterilized) and fruiting phases. Plates were incubated in the dark at 15 °C and colony growth was observed at 21 d.
Isolations from pelotons were categorized as (1) Rhizoctonia-like fungi (slow-growing, monilioid, non-sporulating cream mycelium), (2) other fungi, contaminants or other endophytes (e.g. Trichoderma, Aspergillus, Penicillium, pigmented non-sporulating fungi e.g. Armillaria), (3) bacteria (e.g. Pseudomonas) or (4) dead (failing to grow) (Warcup, 1973). Isolates were labelled in the following order: fungal collection season (L = leafing, B = budding, F = flowering, C = capsule formation, S = senescence, D = dormancy), fungal collection year (0 = 2000, 1 = 2001), fungal isolate (a letter from A to Z for each peloton) and treatment (s = non-surface-sterilized collars), i.e. F1As = one isolate from a flowering plant isolated in 2001 from peloton A, without surface sterilization. Chi-squared tests were conducted in Minitab version 13·1 (Minitab Inc.) with the null hypothesis that the proportion of Rhizoctonia-like fungi obtained was not affected by the fungal collection season from leafing to fruiting or surface-sterilization treatment, at a significance of P ≤ 0·05.
Germination
Seed was naturally pollinated at Meereek State Forest at the plant collection site. Ten mature dehisced seed capsules from different individual plants were collected on 4–18 November 2000. Seeds were removed from capsules and spread out on paper to dry overnight at 15 °C. Seeds were mixed and stored in paper envelopes at 4 °C with silica gel until required. Viability testing was conducted using triphenyltetrazolium chloride (Huynh et al., 2004).
Seed was pre-treated with Tween 80 (BDH) (1 % v/v), shaken vigorously, centrifuged for 1 min at high speed, incubated for 24 h at ambient temperature (22 ± 2 °C), surface-sterilized with 0·5 % NaOCl for 3 min and rinsed with sterile water three times. Single drops containing approx. 50–100 seeds were plated on to a sterile central filter paper (Whatman No. 1) in a 9-cm-diameter Petri dish containing low-nutrient oatmeal agar (2·5 g oatmeal, 12 g sucrose, 0·1 % yeast extract, and 6 g agar in 1 L distilled water, pH 5·3). Simultaneously, the same seed batch was asymbiotically germinated on high nutrient Pa5 (Ramsay et al., 1986) and low nutrient OMA media to test for asymbiotic germination.
Fifty-nine isolates of Rhizoctonia-like fungi (three from 2000 and 56 from 2001) were tested for the ability to induce seed germination. For each isolate, a single 5 mm × 5 mm block of actively growing hyphae was placed next to the seed immediately after plating. For dual inoculations, an extra fungal block was included. An asymbiotic control (no fungus) was included. Each treatment was replicated five times. Plates were incubated in darkness at 15 °C and development assessed 4 weeks later as one of four categories: green leaf formation, prominent (6-fold) swelling, slight (2-fold) swelling or no development. In this study, germination was defined and scored as green leaf formation to reduce false positives or latent pathogens (Warcup, 1981; Ramsay et al., 1986). Germination data did not conform to normality and so were analysed using a non-parametric test, the Kruskal–Wallis (KW) test, in Minitab. Significance was recorded at P ≤ 0·05.
Growth
All seedlings produced were deflasked at 16 weeks and seedlings were maintained in axenic culture on oatmeal agar in 12- to 15-cm-tall glass flasks (250 mL) with breathing vent spots (Elastoplast Hypoallergenic Bandaid spots) at 22 °C with a 14-h photoperiod under four ‘Sylvania’ fluorescent lights (30 µmol m−2 s−1) for 12 months. Leaf length was recorded in seedlings that had survived 12 months after inoculation.
Molecular methods
All 59 Rhizoctonia-like isolates were grown in 20 mL liquid Pa5 medium (Collins and Dixon, 1992) in the dark at 20 °C in sterile 30 mL tissue culture vials (Techno-plas®, Australia) for 10 weeks. A 20–30 mg mycelial aliquot was harvested from each culture and DNA was extracted using either a Qiagen DNeasy® extraction kit (Qiagen, Germany) according to the manufacturer's instructions (except that each sample was ground with a mini-pestle in a 1·5-mL Eppendorf tube with sterile sand) or an alternative method (Gardes and Bruns, 1993). Extracted DNA (total 50 µL for each isolate) was quantified against a GeneRuler™ 100-bp DNA ladder (MBI Fermentas, Germany) by electrophoresis.
Samples containing visible DNA were amplified with the universal primers ITS1 and ITS4 (White et al., 1990), targeting the ITS region of the nuclear ribosomal DNA. Each reaction contained 25 µL in an 0·2-mL thin-walled PCR tube (Axygen Scientific, USA) as follows: 1 µL DNA, 1·5 mm magnesium chloride, 800 µm dNTP mix, 1 µm of each forward and reverse primer, 2·5 µL 10× amplification reaction buffer and 1·1 U Taq polymerase (Biotech®, Australia) made to volume with sterile MilliQ® water. A negative control with water replacing the DNA was included with each set of reactions. Two thermocycling parameters and reaction components were used (Gardes and Bruns, 1993; McLean et al., 1999). The first set of parameters was: denaturation at 94 °C for 8 min; 29 cycles of 94 °C for 1 min, 56 °C for 1 min, and extension at 72 °C for 2 min. The second set of parameters shortened all times: denaturation at 94 °C for 1 s; 34 cycles of 94 °C for 30 s, 55 °C for 30 s, and extension at 72 °C for 60 s. Amplified DNA was separated by electrophoresis as before, photographed over ultraviolet light using a Kodak Digital Science DC120™ camera and product length estimated with Kodak Digital Science ID™ Image Analysis Software by reference to the DNA ladder.
RFLP (restriction fragment length polymorphism) was used on the ITS sequences of the 40 isolates amplified to delimit groups for sequencing. PCR products were digested with one of three endonucleases with a 4-bp recognition site: BsuRI, Hin6I and Taq1 (Fermentas-Progen, Australia). Each reaction contained 10 U of a single enzyme with 5 µL ITS-PCR product, 1 µL 10× buffer and 10 µL sterile MilliQ® water, incubated for 24 h at 37 °C (BsuRI, Hin6I) or 65 °C (TaqI). Restriction digests were separated and visualized as before. Nineteen representative fungi were selected for ITS sequencing: three from 2000 (one leafing, one budding and one flowering) and 16 from 2001 (six leafing, five budding and five flowering). DNA was amplified with primers ITS1 and ITS4 as before and PCR products were purified with a Qiagen Qiaquick® purification kit (Qiagen, Germany) according to the manufacturer's instructions. Sequencing reactions contained 3 µL purified PCR product, 1 µM ABI Prism Big Dye sequencing mix (Version 3·1) containing fluorescent dideoxy-terminator (Applied Biosystems), 0·5 µM of either forward (ITS1) or reverse (ITS4) primer and sterile MilliQ® water to a total volume of 10 µL. PCR protocols for sequencing were as follows: 24 cycles at 96 °C for 30 s, 50 °C for 15 s and 60 °C for 4 min. DNA was purified by sodium acetate precipitation and dried overnight before being sequenced at Micromon (Monash University, Melbourne, Australia) using an ABI 375A automated sequencer (Applied Biosystems).
The 11 full-length sequences obtained were examined using software available through ANGIS (Australian National Genomic Information Service). Fungal sequences were searched in GenBank using Blastn and aligned using Clustal W (Thompson et al., 1994) with the closest matches, Sebacina spp. from Caladenia sources, orchid sources and non-orchid sources. All sequences generated have been deposited in GenBank and all cultures have been deposited at the Royal Botanic Gardens, Melbourne. All sequences were edited manually to the same length (487 bp), comprising ITS1, 5·8S and ITS2 regions with alignment gaps (Felsenstein, 1985), and a neighbor-joining (Saitou and Nei, 1987) bootstrapped dendrogram created with MEGA 4 (Tamura et al., 2007), using the Maximum Composite Likelihood method (Tamura et al., 2004).
RESULTS
Isolation
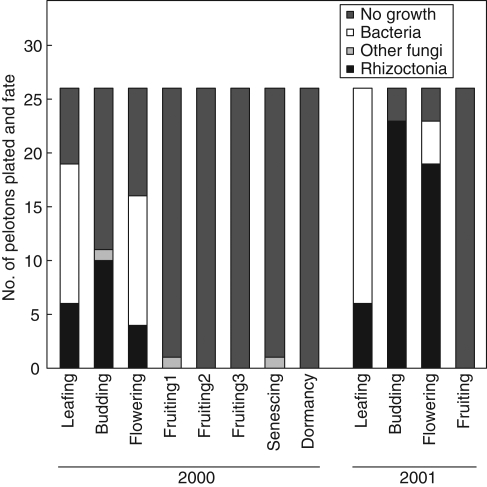
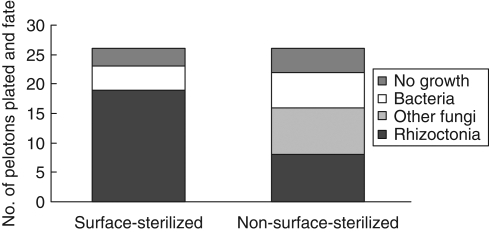
The growth of organisms obtained from pelotons varied with orchid developmental phase (Fig. 1). Rhizoctonia-like fungi were isolated only from leafing–flowering phases and the proportion of pelotons growing from them varied from 15 to 92 %, with greater success in 2001 than 2000. In 2001, the isolation of Rhizoctonia-like isolates was greater than expected at budding and less than expected at fruiting (critical value = 5·99, χ2 Rhizoctonia-like fungi = 36·35, χ2 other categories = 21·8, P < 0·05). Bacteria grew from 11 to 73 % of pelotons but were present only in leafing and flowering phases. The proportion of pelotons without growth generally increased with developmental phase; almost no growth was obtained from pelotons after the flowering phase and this growth comprised common soil-borne fungi such as Trichoderma and Fusarium. In 2001, surface-sterilization of stem-collars at flowering doubled the isolation of Rhizoctonia-like fungi and reduced the isolation of both bacteria and other fungi (critical value = 3·84, χ2 Rhizoctonia-like fungi = 4·48, χ2 other categories = 4·84, P < 0·05; Fig. 2).
Fig. 1.
Total number and proportions of organisms isolated from treated stem-collars at six main developmental host phases of Caladenia formosa. Fruiting 1, 2 and 3 indicates multiple collars used for peloton isolation.
Fig. 2.
Total number and proportions of organisms isolated from treated (surface-sterilized) and untreated (non-surface-sterilized) stem-collars at flowering in 2001.
Germination
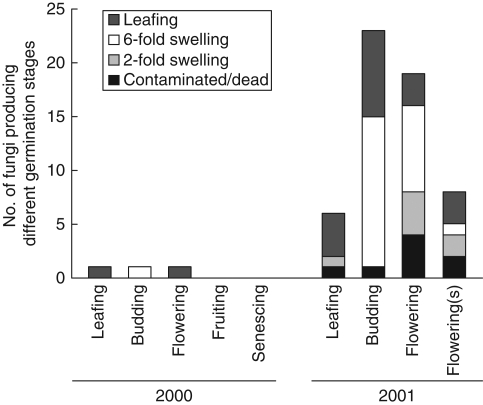
Only one isolate from each phase in 2000 germinated seeds and so data were analysed for 2001 only (Fig. 3). Seed viability using TTC was estimated as low (18·8 % ± 3·8). Only 30 % of isolates induced development to the ‘green leaf’ stage and 42 % to the ‘prominent swelling’ stage, with the remainder showing little or no swelling. Germination to the green leaf stage was affected by orchid developmental phase (KW, P < 0·0001) and reached a maximum of 60 % with isolates from leafing. The number and proportion of isolates that resulted in prominent swelling was greatest in isolates from budding. Isolates giving only slight swelling or no development were found in fruiting and senescing phases. Surface-sterilization of the collar more than doubled the number of isolates that induced green leaf or prominent swelling stages.
Fig. 3.
Stages of development induced by symbiotic germination of Caladenia formosa seeds with Rhizoctonia-like fungi from different orchid developmental phases. Non-surface-sterilized collars denoted by (s).
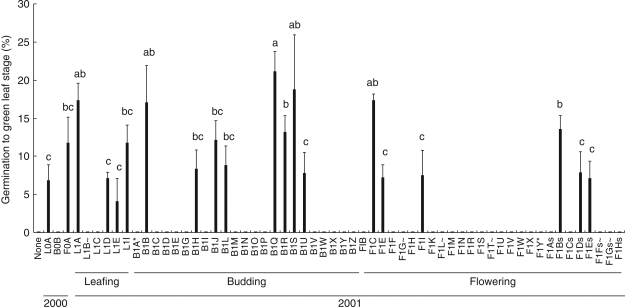
Mean germination (to green leaf stage) ranged from 4 to 21 % (Fig. 4) and there was significant variation among isolates (KW, P < 0·0001). There were, however, large variations within treatments (isolates; coefficient of variation = 11–39 %), even on the same plate. The year of collection influenced germination (KW, P < 0·045) but this may be an artefact as >18 times the number of isolates from 2001 than 2000 were tested. Surface-sterilization influenced the percentage isolation of Rhizoctonia-like fungi but did not influence fungal effectiveness (KW, P > 0·1); only three isolates from each type of treatment germinated seed. Medium also affected the development of asymbiotic seed; on high-nutrient Pa5 medium, germination (to green leaf stage) was >95 % at 12 months, whereas on low-nutrient OMA medium germination was absent and by 12 months seed had only swelled slightly, with no cell division.
Fig. 4.
Percentage germination (mean ± s.e.) of Caladenia formosa at 8 weeks with all Rhizoctonia-like fungal isolates from leafing (L), budding (B) and flowering (F) plants in 2000 and 2001. Explanation of symbols at ends of isolate names: ∼, fungal isolates that were contaminated; *, fungi which germinated seed in previous trials but appeared ineffective in this trial; s, fungi derived from stem-collars that were not surface-sterilized. Means with shared letters are not significantly different (Fisher's l.s.d).
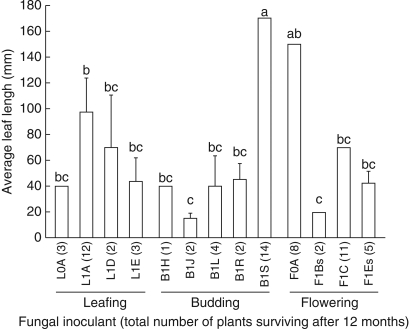
Further growth and survival
Only 69 seedlings, inoculated with 13 of the 20 isolates, survived for 12 months, producing leaves 1–170 mm long (Fig. 5). Survival ranged from 1 to 14 seedlings per isolate and leaf length was affected by the fungal inoculant (KW, P < 0·05). Two isolates (B1S and F0A) induced particularly vigorous growth (leaves >140 mm). Isolates varied widely in uniformity of their effects (coefficient of variation = 0·5–60 %). Neither orchid developmental phase (KW, P > 0·1) nor year of collection (KW, P > 0·1) significantly affected leaf length. Surface-sterilization did not significantly affect seedling growth (KW, P >0·1). Survival and leaf length (Fig. 5) were not related to percentage germination per isolate (Fig. 4). For example, isolates B1Q and B1S had comparably high germination but B1S-inoculated seedlings survived to 12 months and had the longest leaves, whereas B1Q-inoculated seedlings did not. Conversely, some isolates that produced only average germination (e.g. F0A) had relatively high survival and leaf length. Seedling survival and leaf length also varied even in isolates from the same plant (e.g. budding 2001, isolates B1H–B1S).
Fig. 5.
Leaf length (mean ± s.e.) and total number of Caladenia formosa seedlings (in parenthesis) surviving at the time of deflasking in winter 2003 (12 months from inoculation). s = fungal isolates from stem-collars that were untreated. Means with shared letters are not significantly different (Fisher's l.s.d).
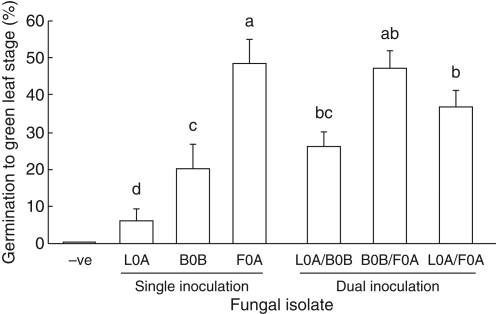
Dual inoculation
Percentage germination varied with isolate for single inocula, with F0A the greatest and L0A the least (Fig. 6). Uninoculated seeds did not swell or develop leaves. There was no significant difference in average germination between single and corresponding dual inoculations of isolates (KW, P > 0·1).
Fig. 6.
Germination (mean ± s.e.) of Caladenia formosa seed with single and co-inoculation of Rhizoctonia-like fungi. Means with shared letters are not significantly different (Fisher's l.s.d).
Molecular methods
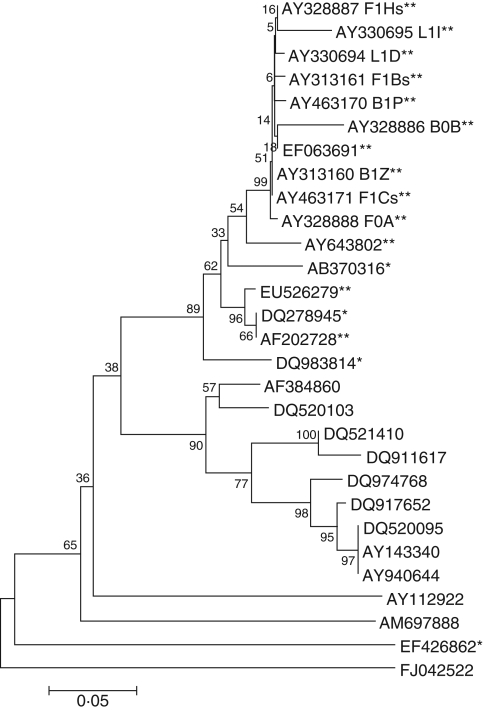
DNA extraction was 75 % efficient and PCR was 89 % efficient, with the result that only 39 of the 59 isolates produced a product of the expected size of approx. 650 bp. All three endonucleases digested the ITS-PCR products and isolates were categorized into four RFLP groups. Most isolates (93 %) were classified as RFLP group I and had identical banding patterns with each of the three enzymes. The remaining isolates appeared to have extra RFLP bands, but on sequencing these proved to be undigested fragments. Of the 19 isolates chosen and sequenced, only 11 produced adequate full-length sequences for analysis. Sequences from two isolates (F1Y and L0A) measured only 227 and 341 bp, respectively, representing the 5·8S ribosomal gene and a partial sequence of ITS2, and were excluded from alignment. The remaining isolates produced sequences of variable lengths (435–510 bp) spanning the ribosomal 5·8S gene, with partial sequences of the ITS1 and ITS2 regions. The sequences of all nine Victorian fungi matched closely with isolates from Caladenia formosa and other Caladenia sources, with similarities ranging from 79 % (L1I and F1Y) to 89 % (L0A) similarity, and were most similar in the 5·8S gene and least similar in the ITS1 region. The neighbor-joining tree (Fig. 7) showed that sequences of the nine Victorian isolates (both effective and ineffective) clustered together on a separate branch within a main branch also containing S. vermifera (AF202728), but separated from it. This main branch was separated from other species of Sebacina from orchid and non-orchid sources. Some isolates from different plants and seasons were identical (i.e. F1Bs and B1P) but exhibited functional differences through seed germination and development (e.g. F1Bs induced leafing stages and B1P induced swelling only).
Fig. 7.
Evolutionary relationships of 29 taxa. The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei, 1987). The bootstrap consensus tree inferred from 1000 replicates (Felsenstein, 1985) is taken to represent the evolutionary history of the taxa analysed (Felsenstein, 1985). Branches corresponding to partitions reproduced in <50 % bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). There was a total of 180 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007). **, Caladenia sources; *, orchid sources; no asterisks, non-orchid sources.
DISCUSSION
This study shows that one plant contains more than one isolate that differs vastly in effectiveness on seed germination and plant growth. Colonization by multiple fungi has been reported from single pelotons (Kristiansen et al., 2001b) and plants (Kristiansen et al., 2001a; McKendrick et al., 2002; Taylor et al., 2003; Kristiansen et al., 2004; McCormick et al., 2004) but their function was untested. Others have tested the mycorrhizal status of the fungi isolated (McCormick et al., 2004) but the efficacy in germination and growth was not quantified.
This study demonstrated the influence of orchid developmental phase of the host on both ease of isolation and effectiveness of mycorrhizal fungi in C. formosa, and suggests that surface sterilization increases the recovery of Rhizoctonia-like fungi to test but the effectiveness varied according to individual isolates. It shows that the fungal isolates that produce the greatest germination may not produce the greatest longer-term orchid growth, suggesting that selection of the ‘best’ isolate for recovery plans should not be made too early and that more than one fungus may be required to optimize both. It also shows that having two fungal isolates during germination, as might be found in the field, has no significant effect on germination and suggests that the presence of genetically similar fungi in one plant in the field may be normal and have longer-term effects.
Molecular comparison of the mycorrhizal fungi in this study suggests that the same fungus can be found colonizing orchids throughout the season. Fungal isolates from C. formosa were similar to S. vermifera, but were both genetically and functionally different, with extreme functional variations from neutralists to highly effective symbionts.
Optimizing isolation
The strong peak in ease of isolation and orchid response to inoculation suggests that collection time can be optimized to improve the number, not only of total isolates, but also of effective isolates collected. The larger proportion of Rhizoctonia-like fungi isolated in early rather than late developmental phases of C. formosa agrees with previous suggestions that mycorrhizal fungi can only be extracted from pre-fruiting phases for C. arenicola (Batty et al., 2001). Specific times for isolation in literature are vague, with only one study (Ramsay et al., 1986) specifying isolation at leafing phases and only two studies on bacterial localization specifying winter or when moisture is maximal (Wilkinson et al., 1989; Tsavkelova et al., 2003). The lack of mycorrhizal fungus growth from pelotons in later developmental phases is probably due to the increasing collapse and death of the fungi forming the pelotons (Huynh et al., 2004). This collapse may be due to starvation of the symbiotic endophyte in the face of growing competition from the developing fruit for photosynthates, as in the legume–rhizobia symbiosis (Lawrie and Wheeler, 1974) or merely leaf senescence preceding dormancy. The poorer isolation of mycorrhizal fungi in 2000 than 2001 may be linked to the thick hyphae inside the cells of the 2000 collars only (Huynh et al., 2004), possibly a non-symbiont or pathogen that out-competes the mycorrhizal fungus for nutrients. This may also explain the poorer effectiveness of isolates from comparable phases from 2000 than 2001.
Surface-sterilization improved the isolation of effective mycorrhizal fungi, suggesting that surface-sterilization essentially removed fungi and bacteria on the collar surface, as the fungi from untreated collars were common soil genera such as Fusarium and Trichoderma. It is possible to isolate effective mycorrhizal fungi without surface-sterilization (Clements, 1981; Dijk and Eck, 1995; Bonnardeaux et al., 2007), but this study has shown that it is desirable in order to optimize the number of potentially effective fungi isolated.
Germination versus growth
Finding the most effective fungus to produce both the greatest germination and the greatest growth remains difficult and unpredictable. The lack of correspondence between germination and seedling development suggests that fungi from C. formosa producing the greatest germination do not necessarily produce the greatest growth, as shown in northern hemisphere orchids by Rasmussen (1995). Few studies have compared germination with post-leafing phases, and fewer have observed deflasking proportions (Rubluo et al., 1989; Bohm, 1991; Ramsay and Stewart, 1998; Ortiz-Barney and Ackerman, 1999; Stewart and Zettler, 2002). Even fewer studies have measured longer-term survival and regeneration in situ (Batty et al., 2006).
Molecular data in this study suggest that each collar has several genetically closely related but not identical mycorrhizal fungi, all of which may be required for optimal survival and growth. Some authors (McKendrick et al., 2000; Kristiansen et al., 2001b; Dearnaley, 2007) suggest that in nature, some orchids exist with multiple mycorrhizal fungi and Kristiansen et al. (2001b) have shown that genetically different fungi can co-exist even within one peloton. Sharma et al. (2003) implied that different fungi were required for different developmental phases. The functional differences observed in this study advocate the requirement for other researchers to also confirm the connection between fungal associates with their function in other orchids. This will lead to more accurate conclusions on fungal relationships and whether a different group of mycorrhizal associates also express different functional roles.
The lack of evidence for one fungus having a clear selective advantage in both germination and growth has important practical consequences, as the routine use of only one fungus as inoculum for ex-situ growth and plant translocation in recovery plans may eliminate other vital fungi from the orchid mycorrhizal population and hamper conservation efforts. Requirements for multiple fungi would have significant implications and could necessitate artificial inoculations at existing re-introduction sites or for depleted populations in situ.
Although direct seed germination testing is time-consuming and dependent on stringent conditions, there is currently no other selection criterion that can be used to select highly effective fungi.
Variation in mycorrhizal fungi
Caladenia formosa appears to be mycorrhizal with several genetically similar fungi forming one group that is most similar but not identical to S. vermifera in the rDNA ITS region. Fungal isolates with identical sequences were found from different plants, developmental phases and years, suggesting that the same fungus is retained in either the old tuber or the surrounding soil for re-colonization in subsequent seasons (Warcup, 1971, 1981; Bohm, 1991). The loss of growth and sequencing information from >33 % of isolates probably underestimates the true diversity of mycorrhizal fungi. Difficulties in growth of fungi associated with Caladenia sp. have been reported previously (Ramsay et al., 1986) and commonly occur with other orchid fungi in culture (Zelmer and Currah, 1995) with similar ‘sebacinoids’ from epacrids completely failing to grow (Allen et al., 2003). Direct amplification of pelotons has been used by others to bypass cultural growth difficulties (Kristiansen et al., 2001b; Allen et al., 2003) and this may be a better strategy, but is only feasible if DNA can be separated by base composition (Kristiansen et al., 2001b) and its mycorrhizal effectiveness confirmed.
Fungi from C. formosa are phylogenetically most similar to the only ITS sequence available from S. vermifera, as expected from previous studies (Warcup, 1971; Clements, 1981; Warcup, 1981; Ramsay et al., 1986), but are not the same identity. The precise homology defining a species using phylogenetic evidence is speculative (Taylor et al., 2000) but rDNA ITS sequences are considered likely to be from the same species if they are 98–100 % similar (Sreenivasaprasad et al., 1996; McLean et al., 1999). Phylogenetic similarities below this limit (particularly in an ITS region that is rapidly evolving) are more likely to be in the same genus or family rather than species. The 79–89 % homology of C. formosa isolates to S. vermifera in this study suggests that they are probably not S. vermifera but are within Sebacina and the Sebacinales.
Species recognition using molecular evidence provides an alternative method for the identification of asexual and morphologically identical orchid fungi, but identification to species is still reliant on the production of teleomorphic stages, which have not been produced since the studies of Warcup and Talbot (1967). Re-examination and sequencing of such reference cultures could potentially improve our knowledge of the identity of mycorrhizal isolates.
Variation among isolates in effectiveness
Genetic similarity in the rDNA ITS region was not a predictor of effectiveness (the ability to germinate C. formosa seeds and support their further growth and development), as effectiveness varied even in the five fungi genetically identical in this region. Morphologically identical mycorrhizal fungi, even from the same plant, varied widely in effectiveness. These results emphasize the necessity of conducting trials with numerous isolates from even one plant and it may be the combination of multiple fungi that leads to an effective symbiosis.
Considerable functional and genomic differences of mycorrhizal fungi even from the same plant, as shown in this study, suggest that the diversity of fungi may be greater than previously recognized. This means that for the continuation of important fungal–host associations, more efforts are needed to preserve fungal diversity rather than systematically testing and storing single isolates.
ACKNOWLEDGMENTS
We thank Maggie Nightingale and Mark Clements for fungal cultures. We thank Ben Kefford, Fiona Coates and three anonymous reviewers for helpful critiques on the manuscript. James Todd, Andrew Pritchard and An Huynh provided assistance with field work. Plants were collected under permits 10001562 and 10000969 from the Department of Sustainability and Environment Victoria. We dedicate this to Cassandra McLean, may you rest in peace. This study was supported by the University of Melbourne MRS Scholarship Grant.
LITERATURE CITED
- Allen TR, Millar T, Berch SM, Berbee ML. Culturing and direct DNA extraction find different fungi from the same ericoid mycorrhizal roots. New Phytologist. 2003;160:255–272. doi: 10.1046/j.1469-8137.2003.00885.x. [DOI] [PubMed] [Google Scholar]
- Batty AL, Dixon KW, Brundrett M, Sivasithamparam K. Long-term storage of mycorrhizal fungi and seed as a tool for the conservation of endangered Western Australian terrestrial orchids. Australian Journal of Botany. 2001;49:619–628. [Google Scholar]
- Batty AL, Brundrett MC, Dixon KW, Sivasithamparam K. In situ symbiotic seed germination and propagation of terrestrial orchid seedlings for establishment at field sites. Australian Journal of Botany. 2006;54:275–381. [Google Scholar]
- Bohm J. In vitro propagation of terrestrial orchids: Diuris emarginata R. Br. The Orchadian. 1991;10:110–113. [Google Scholar]
- Bonnardeaux Y, Brundrett M, Batty A, Dixon K, Koch J, Sivasithamparam K. Diversity of mycorrhizal fungi of terrestrial orchids: compatibility webs, brief encounters, lasting relationships and alien invasions. Mycological Research. 2007;111:51–61. doi: 10.1016/j.mycres.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Bougoure JJ, Bougoure DS, Cairney JW, Dearnaley JD. ITS-RFLP and sequence analysis of endophytes from Acianthus, Caladenia and Pterostylis (Orchidaceae) in southeastern Queensland. Mycological Research. 2005;109:452–460. doi: 10.1017/s095375620500225x. [DOI] [PubMed] [Google Scholar]
- Cameron DD, Leake JR, Read DJ. Mutualistic mycorrhiza in orchids: evidence from plant-fungus carbon and nitrogen transfers in the green-leaved terrestrial orchid Goodyera repens. New Phytologist. 2006;171:405–16. doi: 10.1111/j.1469-8137.2006.01767.x. [DOI] [PubMed] [Google Scholar]
- Cameron DD, Johnson I, Leake JR, Read DJ. Mycorrhizal acquisition of inorganic phosphorus by the green-leaved terrestrial orchid Goodyera repens. Annals of Botany. 2007;99:831–834. doi: 10.1093/aob/mcm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements MA. Developments in the symbiotic germination of Australian terrestrial orchids. In: Stewart J, van der Merwe CN, editors. Proceedings from the 10th World Orchid Conference; Durban. Johannesburg: South African Orchid Council; 1981. pp. 269–273. [Google Scholar]
- Collins MT, Dixon KW. Micropropagation of an Australian terrestrial orchid Diuris longifolia R. Br. Australian Journal of Experimental Agriculture. 1992;32:131–135. [Google Scholar]
- Currah RS, Smreciu EA, Hambleton S. Mycorrhizae and mycorrhizal fungi of boreal species of Platanthera and Coeloglossum (Orchidaceae) Canadian Journal of Botany. 1990;68:1171–1181. [Google Scholar]
- Dearnaley JD. Further advances in orchid mycorrhizal research. Mycorrhiza. 2007;17:475–486. doi: 10.1007/s00572-007-0138-1. [DOI] [PubMed] [Google Scholar]
- Dijk E, Eck ND. Effects of mycorrhizal fungi on in vitro nitrogen response of some Dutch indigenous orchid species. Canadian Journal of Botany. 1995;73:1203–1211. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. ITS primers with enhanced specificity for Basidiomycetes: application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Gebauer G, Meyer M. 15N and 13C natural abundance of autotrophic and myco-heterotrophic orchids provides insight into nitrogen and carbon gain from fungal association. New Phytologist. 2003;160:209–223. doi: 10.1046/j.1469-8137.2003.00872.x. [DOI] [PubMed] [Google Scholar]
- Huynh TT, McLean CB, Coates F, Lawrie AC. Effect of developmental stage and peloton morphology on success in isolation of mycorrhizal fungi in Caladenia formosa (Orchidaceae) Australian Journal of Botany. 2004;52:231–241. [Google Scholar]
- Julou T, Burghardt B, Gebauer G, Berveiller D, Damesin C, Selosse MA. Mixotrophy in orchids: insights from a comparative study of green individuals and nonphotosynthetic individuals of Cephalanthera damasonium. New Phytologist. 2005;166:639–653. doi: 10.1111/j.1469-8137.2005.01364.x. [DOI] [PubMed] [Google Scholar]
- Kristiansen KA, Rasmussen FN, Rasmussen HN. Seedlings of Neuwiedia (Orchidaceae subfamily Apostasioideae) have typical orchidaceous mycotrophic protocorms. American Journal of Botany. 2001;a 88:956–959. [PubMed] [Google Scholar]
- Kristiansen KA, Taylor DL, Kjøller R, Rasmussen HN, Rosendahl D. Identification of mycorrhizal fungi from single pelotons of Dactylorhiza majalis (Orchidaceae) using single-strand conformation polymorphism and mitochondrial ribosomal large subunit DNA sequences. Molecular Ecology. 2001;b 10:2089–2093. doi: 10.1046/j.0962-1083.2001.01324.x. [DOI] [PubMed] [Google Scholar]
- Kristiansen KA, Freudenstein JV, Rasmussen FN, Rasmussen HN. Molecular identification of mycorrhizal fungi in Neuwiedia veratrifolia (Orchidaceae) Molecular Phylogenetics and Evolution. 2004;33:251–258. doi: 10.1016/j.ympev.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Lawrie AC, Wheeler CT. The effects of flowering and fruit formation on the supply of photosynthetic assimilates to the nodules of Pisum sativum L. in relation to the fixation of nitrogen. New Phytologist. 1974;73:1119–1127. [Google Scholar]
- Leake JR. Plants parasitic on fungi: unearthing the fungi in myco-heterotrophs and debunking the ‘saprophytic’ plant myth. Mycologist. 2005;19:113–122. [Google Scholar]
- McCormick MK, Whigham DF, O'Neill J. Mycorrhizal diversity in photosynthetic terrestrial orchids. New Phytologist. 2004;163:425–438. doi: 10.1111/j.1469-8137.2004.01114.x. [DOI] [PubMed] [Google Scholar]
- McKendrick SL, Leake JR, Taylor DL, Read DJ. Symbiotic germination and development of myco-heterotrophic plants in nature: ontogeny of Corallorhiza trifida and characteristics of its mycorrhizal fungi. New Phytologist. 2000;145:523–537. doi: 10.1046/j.1469-8137.2000.00603.x. [DOI] [PubMed] [Google Scholar]
- McKendrick SL, Leake JR, Taylor DL, Read DJ. Symbiotic germination and development of the myco-heterotrophic orchid Neottia nidus-avis in nature and its requirement for locally distributed Sebacina spp. New Phytologist. 2002;154:233–247. [Google Scholar]
- McLean CB, Cunnington JH, Lawrie AC. Molecular diversity within and between ericoid endophytes from the Ericaceae and Epacridaceae. New Phytologist. 1999;144:351–358. [Google Scholar]
- Ortiz-Barney E, Ackerman JD. The cost of selfing in Encyclia cochleata (Orchidaceae) Plant Systematics and Evolution. 1999;219:55–64. [Google Scholar]
- Pope EJ, Carter DA. Phylogenetic placement and host specificity of mycorrhizal isolates belonging to AG-6 and AG-12 in the Rhizoctonia solani species complex. Mycologia. 2001;93:712–719. [Google Scholar]
- Raleigh RE, Cross RG, Lawrie AC, Coates F, Moorrees ACA. Research into the propagation of eastern Australian Caladenia. In: Barrett RL, Dixon KW, editors. Book of Extended Abstracts. Proceedings from the First International Orchid Conservation Congress; September 24–28, 2001; Perth, Western Australia. Perth: Plant Conservation Association Inc.; 2001. p. 115. [Google Scholar]
- Ramsay MM, Stewart J. Re-establishment of the lady's slipper orchid (Cypripedium calceolus L.) in Britain. Botanical Journal of the Linnean Society. 1998;126:173–181. [Google Scholar]
- Ramsay RR, Dixon KW, Sivasithamparam K. Patterns of infection and endophytes associated with Western Australian orchids. Lindleyana. 1986;1:203–214. [Google Scholar]
- Rasmussen HN. Terrestrial orchids from seed to mycotrophic plant. Cambridge: Cambridge University Press; 1995. [Google Scholar]
- Rubluo A, Chavez V, Martinez A. In vitro seed germination and re-introduction of Bletia urbana (Orchidaceae) in its natural habitat. Lindleyana. 1989;4:68–73. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sharma J, Zettler LW, van Sambeck JW, Ellersieck MR, Starbuck CJ. Symbiotic seed germination and mycorrhizae of federally threatened Platanthera praeclara. American Midland Naturalist. 2003;149:104–120. [Google Scholar]
- Sreenivasaprasad S, Mills PR, Meehan BM, Brown AE. Phylogeny and systematics of 18 Colletotrichum species based on ribosomal DNA spacer sequences. Genome. 1996;39:499–512. doi: 10.1139/g96-064. [DOI] [PubMed] [Google Scholar]
- Stewart SL, Zettler LW. Symbiotic germination of three semi-aquatic rein orchids (Habeniaria repens, H.quinquiseta, H. macroceratitis) from Florida. Aquatic Botany. 2002;72:25–35. [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences of the USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4·0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Taylor DL, Bruns TD, Szaro TM, Hodges SA. Divergence in mycorrhizal specialization within Hexalectris spicata (Orchidaceae), a nonphotosynthetic desert orchid. American Journal of Botany. 2003;90:1168–1179. doi: 10.3732/ajb.90.8.1168. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DL, Kroken S, et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsavkelova EA, Lobakova ES, Kolomeitseva GL, Cherdyntseva TA, Netrusov AI. Localization of associative Cyanobacteria on the roots of epiphytic orchids. Microbiology. 2003;72:86–91. [PubMed] [Google Scholar]
- Wang B, Qiu YL. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza. 2006;16:299–363. doi: 10.1007/s00572-005-0033-6. [DOI] [PubMed] [Google Scholar]
- Warcup JH. Specificity of mycorrhizal association in some Australian terrestrial orchids. New Phytologist. 1971;70:41–46. [Google Scholar]
- Warcup JH. Symbiotic germination of some Australian terrestrial orchids. New Phytologist. 1973;72:387–392. [Google Scholar]
- Warcup JH. The mycorrhizal relationships of Australian orchids. New Phytologist. 1981;87:371–381. [Google Scholar]
- Warcup JH, Talbot PHB. Perfect states of Rhizoctonias associated with orchids. New Phytologist. 1967;66:631–641. [Google Scholar]
- Waterman RJ, Bidartondo MI. Deception above, deception below: linking pollination and mycorrhizal biology of orchids. Journal of Experimental Botany. 2008;59:1085–1096. doi: 10.1093/jxb/erm366. [DOI] [PubMed] [Google Scholar]
- White JJ, Bruns TD, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Gelfand MA, Sninsky JJ, White JJ, editors. PCR protocols – a guide to methods and applications. New York, NY: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Wilkinson KG, Dixon KW, Sivasithamparam K. Interaction of soil bacteria, mycorrhizal fungi and orchid seed in relation to germination of Australian orchids. New Phytologist. 1989;112:429–435. [Google Scholar]
- Zelmer CD, Currah RS. Evidence for a fungal liaison between Corallorhiza trifida (Orchidaceae) and Pinus contorta (Pinaceae) Canadian Journal of Botany. 1995;73:862–866. [Google Scholar]