Figure 6. Links between the DNA methylation and piRNA pathways.
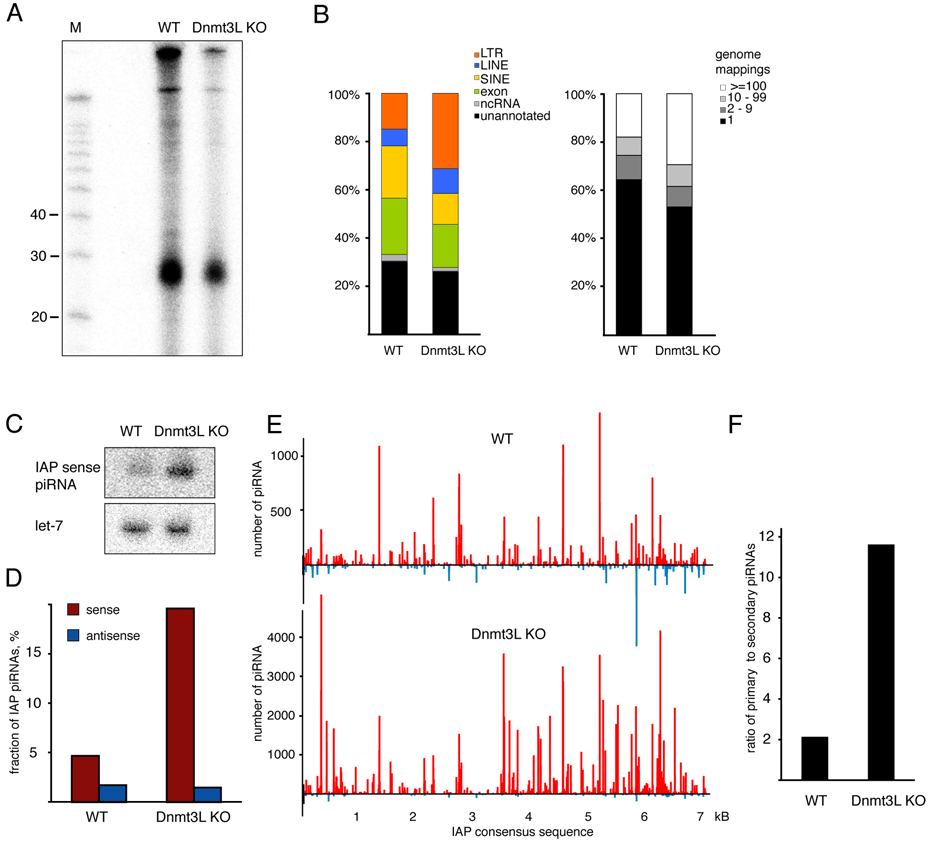
(A) MILI-piRNA complexes were immunoprecipitated from testes of 10 dpp wild type and Dnmt3L knock-out animals, and isolated RNAs were 5’ labeled. (B) piRNA populations were analyzed in Dnmt3L mutants. piRNAs isolated from MILI complexes shown in (A) were cloned, sequenced and annotated. Shown are the annotation (left panel) and genomic mapping (right panel) for RNAs from wild-type and Dnmt3L mutants. (C) Northern hybridization with LNA probe to detect an abundant IAP sense piRNA in 10 dpp testes of wild type and Dnmt3L KO mice. Hybridization with let-7 miRNA was used as a loading control. (D) The fraction of sense and antisense IAP piRNAs is shown for wild-type and Dnmt3L mutant libraries. (E) The distribution of piRNAs on the IAP retrotransposon consensus is shown for MILI complexes from wild-type and Dnmt3L mutant animals. (F) Primary and secondary IAP piRNA in Dnmt3L mutants. Shown is the ratio of primary (1U, no-10A) to secondary (no-1U, 10A) piRNAs in the indicated libraries.