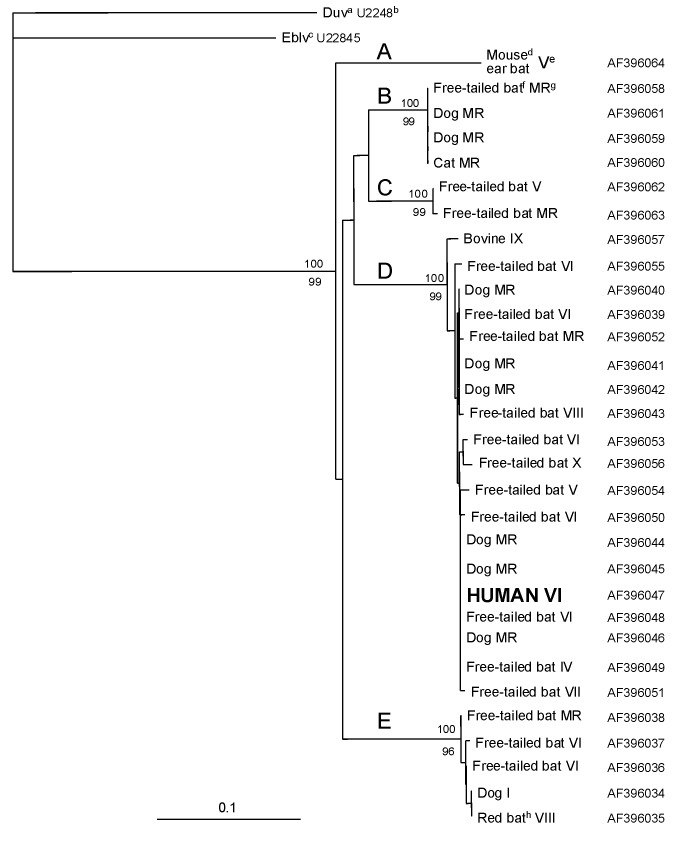
Figure 2.
Neighbor-joining tree comparing the human rabies isolate with representatives of the rabies genetic variants obtained from insectivorous bats and domestic animals in Chile (18). Bootstrap values obtained from 100 resamplings of the data by using distance matrix (top) and parsimony methods (bottom) are shown at nodes corresponding to the lineages representing the rabies virus variants (A, B, C, D, and E) currently circulating in Chile. Only bootstrap values >50% are shown at the branching points. The bar at the left corner indicates 0.1 nucleotide substitutions per site. aDuvenhage virus, bGenBank accession number, cEuropean bat Lyssavirus, dMyotis chiloensis, eRoman numerals indicate the administrative region where the sample was obtained, fTadarida brasiliensis, gMetropolitan region, hLasiurus borealis.