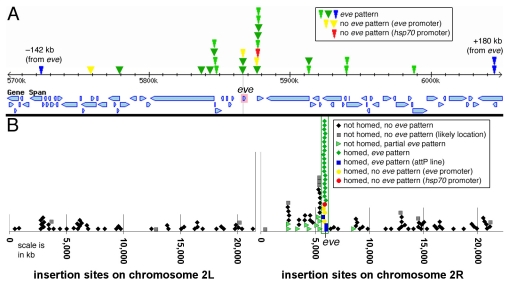
Fig. 5.
Insertion sites of transgenes carrying the insulator/homing region. (A) Genetic map covering 350 kb of chromosome 2R, from 5700 to 6050 kb. The transcription start site of eve (at 5867 kb on this scale) is marked with a vertical gray line. Transcription units (as seen in FlyBase) are shown as blue boxes. Insertion sites of homed lines are indicated by narrow arrowheads for eZ and hsp70-lacZ constructs (above the dotted line in Fig. 3), and wide arrowheads for eZAR-MeW constructs (below the dotted line in Fig. 3). Colors indicate the following: green, transgenes expressing β-gal in an essentially complete eve pattern; blue, attP RR11K insertions that also show an eve-like pattern; red, an SR105 transgene with an hsp70-lacZ reporter not expressed in an eve-like pattern; yellow (near the red arrowhead), CR105 with an eve-promoter-lacZ reporter that gave expression only in the AR; yellow (just upstream of the eve start site), an attP-RR11K insertion that does not give an eve-like β-gal expression. (B) Distribution of insertion sites of eZ and eZAR-MeW transgenes on the second chromosome. The region shown in A is bracketed by two vertical green lines. Note that insertion sites are concentrated near the eve locus, and transgenes that communicate with endogenous eve (green arrowheads) are up to 3300 kb away. Most other insertion lines within this region (all but three of the black diamonds) carry the AR and mesodermal eve enhancers (eZAR-MeW), obscuring potential long-range interactions (see text). Gray squares correspond to the most likely locations of transgenes where inverse PCR-derived sequences gave somewhat ambiguous results (see Table S1 in the supplementary material).