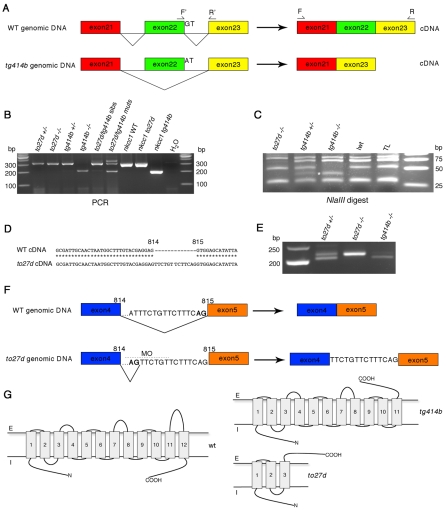
Fig. 5.
Genomic sequencing reveals nkcc1 splice site mutations in both lte alleles. (A) ltetg414b is a G→A transition mutation in the splice donor site of exon 22. The resulting transcript lacks 112 bp and is shifted out of frame. (B) Amplification of nkcc1 cDNA with primers flanking the lost exon (see A; F, forward, R, reverse) reveals a 203 bp band in tg414b homozygotes, compound heterozygotes and full-length subcloned tg414b mutant cDNA, as compared with a 315 bp band amplified from the to27d mutant and sibling embryos, full-length wild-type (WT) and to27d subcloned mutant cDNA. (C) NlaIII digest of PCR products generated with primers flanking the lesion in genomic DNA reveals the introduction of a novel NlaIII site in tg414b mutants, cleaving a 53 bp fragment into 40 bp (asterisk) and 13 bp fragments. (D,E) Sequencing of cDNA from the to27d allele reveals a 13 bp insertion between bases 814 and 815 (D), which can be seen in an RT-PCR reaction using primers flanking this sequence (E): a band of 216 bp is present in the to27d line, as compared with the wild-type 203 bp band, amplified here from the tg414b line. (F) Analysis of to27d genomic DNA reveals a T→G transversion mutation in intron 4, introducing a novel splice site 13 bp upstream of the wild-type site. MO indicates the position of the splice-blocking morpholino used in the rescue experiment (see Fig. 6). (G) Theoretical protein topology models (Tusnàdy and Simon, 1998; Tusnàdy and Simon, 2001). The wild-type protein has 12 transmembrane helices; both the N- and C-termini are predicted to be intracellular. The tg414b protein would be truncated after the eleventh transmembrane helix and lose its C-terminal intracellular localisation. In to27d, a severely truncated isoform is predicted, with only three transmembrane helices remaining. E, extracellular; I, intracellular.