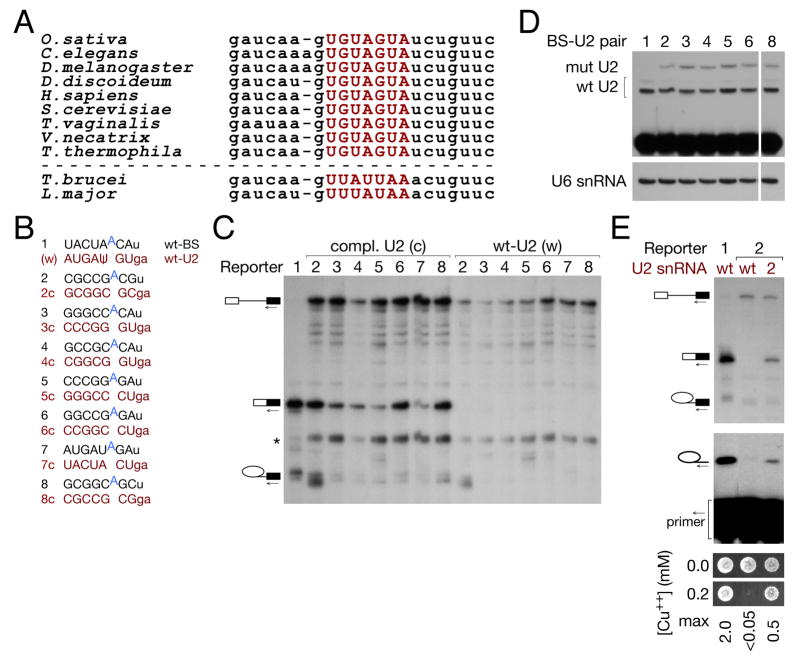
Figure 3. A wide variety of grossly substituted BS-U2 pairs are functional for splicing, and are orthogonal to the endogenous splicing machinery.
(A) Alignment of U2 snRNA sequences from (S. cerevisiae) positions 26 to 49 from diverse organisms: the BS-binding region, highlighted in red, is 100% conserved in crown group eukaryotes.
(B) Schematic of wt and grossly substituted reporter and U2 constructs used in (C).
(C) Primer extension with a 3′ exon primer indicates that grossly substituted branch sites allow splicing catalysis in the presence of their cognate U2 (c) [numbers refer to (B)], but not wt U2 (w).
(D) Primer extension analysis with ddTTP replacing dTTP to assay the expression of U2 snRNAs with wild-type and GC-substituted (orthogonal) BS-binding regions.
(E) Primer extension on total RNA from the indicated strains with a BS-proximal intronic primer (middle panel) and copper growth (lower panel) indicate that the expected branch nucleotide is used and that the mRNA resulting from splicing is functional, for representative grossly substituted BS-U2 duplex 2 from (B).