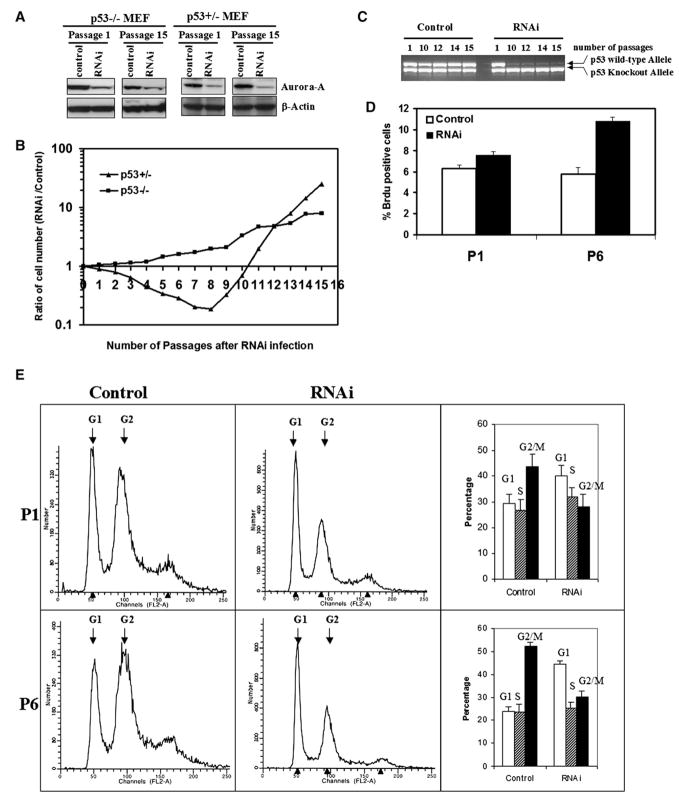
Figure 4. Aurora-A Downregulation by RNAi in p53 Heterozygous and Null MEFs.
Two controls for the experiment were pSUPER vector without RNAi or with scrambled RNAi (see Experimental Procedures), both of which gave the same results.
(A) Aurora-A protein levels are downregulated by RNAi in p53−/− MEFs after selection with 2 μg/ml puromycin.
(B) Downregulation of Aurora-A in p53+/− MEFS produces a clear growth disadvantage at early passages, but growth advantage at late passages, compared with their untransfected counterparts. p53 null MEFs transfected with the same RNAi construct showed similar behavior independently of the presence of Aurora-A RNAi at early passages, and a clear growth advantage at late passages.
(C) Loss of the remaining p53 allele in p53+/− MEFs treated with Aurora-A RNAi. p53+/− cells in the presence of scrambled (control) RNAi maintain the normal p53 allele until around passage 25, but p53+/− cells treated with Aurora-A RNAi lose the wild-type p53 allele between passages 5 and 10.
(D) Percentage of BrdU-positive cells in p53−/− cultures treated with Aurora-A RNAi (right column) compared to controls (left column). Experiments were carried out with cells at passages 1 and 6. Inhibition of Aurora-A leads to a net increase in the number of proliferating cells. This experiment was repeated three times and the results are statistically significant (*p < 0.01). Error bars stand for mean ± SD of three different experiments.
(E) FACS profiles of p53−/− cells in the presence or absence of Aurora-A RNAi. The same cell populations used for the analyses shown in (D) were analyzed by flow cytometry. The proportion of cells in the G2/M phases of the cell cycle was substantially reduced, suggesting, when taken together with the BrdU incorporation assay shown in (D), that inhibition of Aurora-A releases a G2/M checkpoint and stimulates cell cycle progression. Error bars stand for mean ± SD of three different experiments.