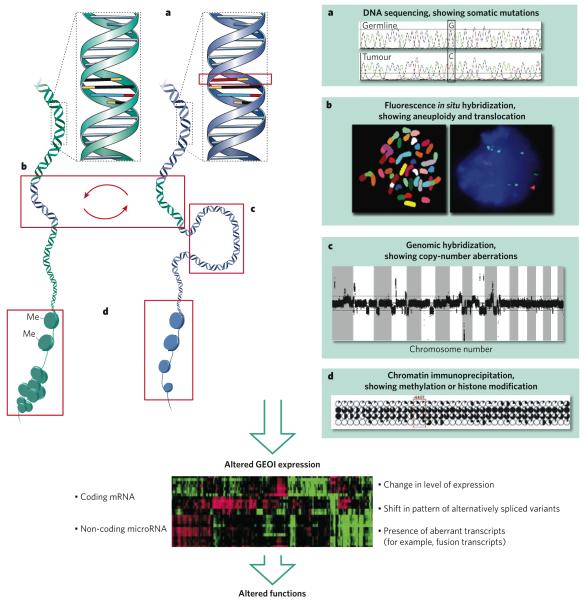
Figure 1. Various types of genomic and epigenomic aberration in cancers.
The main types of genomic and epigenomic aberration are illustrated together with examples of how they can be detected. a, Changes in DNA sequence, such as point mutations, can be assessed by DNA-sequencing techniques. b, Changes in genomic organization can be assessed by using fluorescence in situ hybridization. In the example shown, DNA segments are exchanged between the two (blue and green) DNA molecules. c, Changes in DNA copy number, such as those that result from amplification, can be assessed by using comparative genomic hybridization. d, Changes in DNA methylation and the resultant changes in chromatin structure can be assessed by using chromatin immunoprecipitation plus microarray analysis of immunoprecipitated DNA. Each of these types of change can alter the expression levels of genes or non-coding microRNAs (referred to here as genetic elements of interest, GEOIs), alter the splicing patterns of transcripts, or change gene function through mutation or through creating chimaeric genes. Many of these events can be as assessed by microarray analysis. These changes ultimately translate into altered functions, leading to a diseased state, such as cancer.