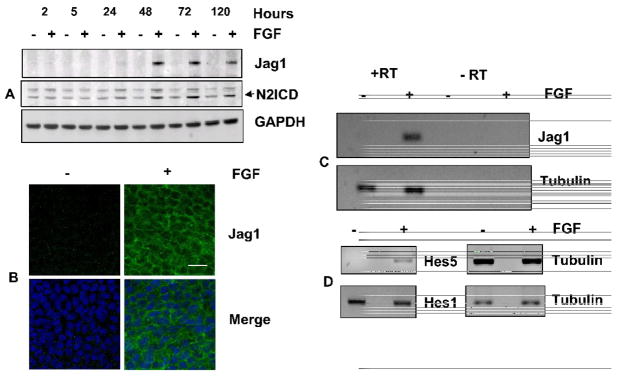
Fig. 3. FGF induces Jag1 and activates Notch signaling.
(A) Representative immunoblot of Jag1 and N2ICD from lens epithelial explants exposed to 100ng/mL FGF (+) or 0.1% BSA (−) for indicated times (hours). GAPDH has been used as a loading control. The two bands recognized by the N2ICD antibody likely represent sequential cleavage products of Notch2. Lower band represents the N2ICD (B) Representative immunofluorescence of Jag1 in the central region of rat lens epithelia cultured in the presence or absence of FGF for 4 days shows that FGF induces uniform expression of Jag1 in all cells in this region and demonstrates that Jag1(green) is localized along cell-cell boundaries. Nuclei have been stained with DAPI (blue). Scale bar is applicable to all images of panel B and represents 20 μm length (C) FGF induction of Jag1 mRNA was demonstrated by RT-PCR using total RNA isolated from CE explants cultured in the presence or absence of FGF (100 ng/mL) for 24 hours. Tubulin RT-PCR was performed as a positive control. Reactions lacking reverse transcriptase (RT) served as negative controls. Analyses of at least three different replicates provided similar results. (D) The ability of FGF to activate Notch signaling was examined by RT-PCR for Notch effectors Hes5 and Hes1, using total RNA isolated from CE explants cultured in the presence or absence of FGF (100 ng/mL) for 24 hours. Tubulin RT-PCR was performed as a positive control. Negative controls lacking reverse transcriptase (RT) showed no amplified products (not shown). Analyses of at least three different replicates provided similar results.