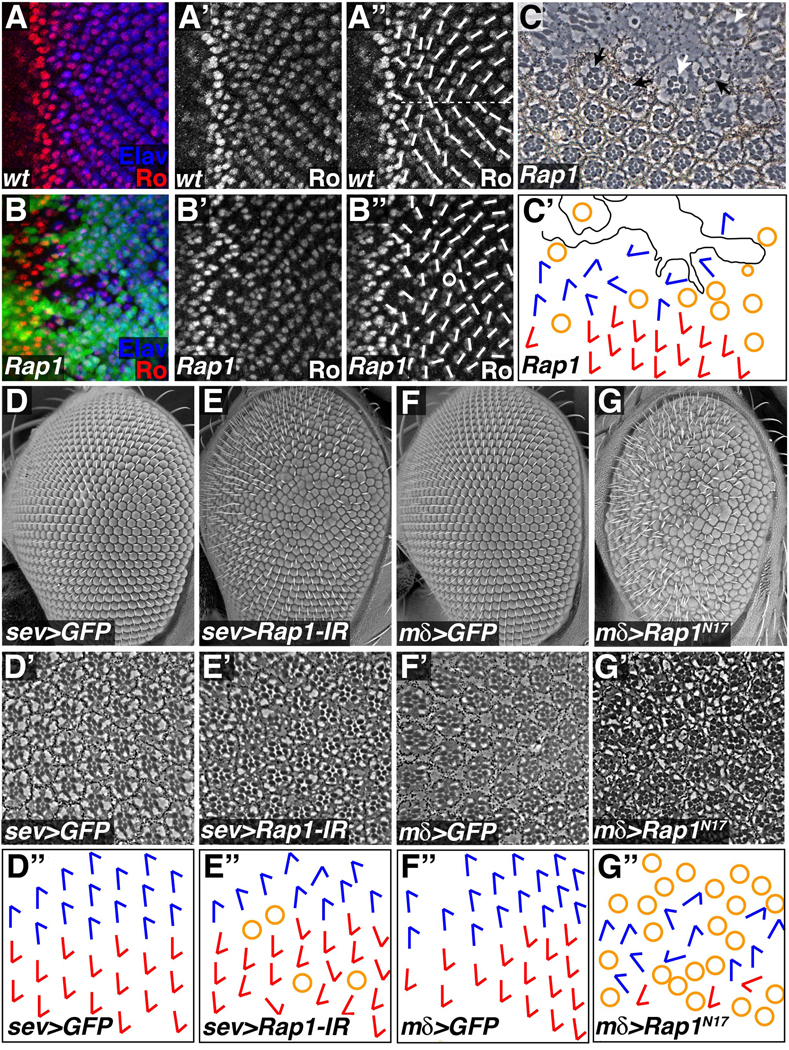
Figure 5. Rap1 affects ommatidial rotation in the developing eye.
(A,B) Eye imaginal discs were stained for Rough (Ro) and Elav. Short white lines indicate ommatidial orientation, while the horizontal dotted line marks the equator. (A) In wildtype discs a regular pattern of ommatidial orientation is apparent. (B,C) Clones of cells lacking Rap1 (GFP negative) were generated using the Flp/FRT system. (B) Rap1 mutant cells disrupt the overall pattern of ommatidial rotation. Circle denotes an ommatidium with abnormal numbers of photoreceptors. (C) In tangential sections of adult eyes Rap1 mutant cells lack pigment. Scar-like tissue often formed in Rap1 mutant clones. At the edges of clones misaligned ommatidia containing Rap1 mutant cells (white arrow), or entirely wildtype cells (black arrows) could be found. White arrowhead indicates an ommatidium containing Rap1 mutant cells that has a normal alignment. (C´) Schematic representation of the section in C is shown. Blue and red arrows indicate ommatidial orientation; yellow circles indicate incomplete ommatidia; black line demarcates scar tissue. (D–G) Loss and gain of Rap1 function in subsets of cells also disrupts ommatidial rotation. SEM micrographs (D–G), tangential sections (D´–G´), and schematic representations of ommatidial orientation (D´´–G´´) are shown for each genotype. (E) sev-Gal4 in combination with Rap1-IR was used to knock down Rap1 function in a subset of differentiating photoreceptors (R3, R4, R7) and cone cells. (G) Similarly, mδ0.5-Gal4 was used to express Rap1N17 in R4 and weakly in R3 and R7. In both cases, reduction of Rap1 activity significantly increased ommatidial misalignment when compared to controls (D and F respectively).