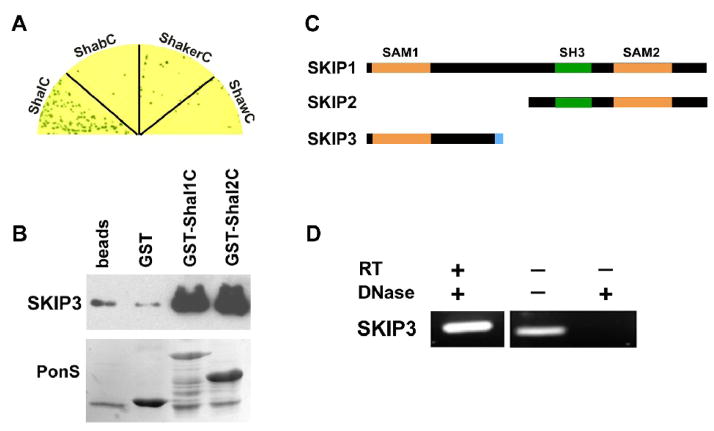
Fig. 1. The Novel Protein, SKIP3, Interacts with the C-terminus of Shal K+ Channels.
(A) SKIP3 was tested in direct Y2H tests with the C-termini of representative isoforms of voltage-gated K+ channels from the four major subfamilies, including Shal1 (ShalC), ShA1 (ShakerC), Shab-RB (ShabC), and Shaw2 (ShawC). Shown are co-transfected yeast grown on stringent media and X-α-Gal to select for interaction. Note that growth and a blue color, indicating interaction, is specific for ShalC. (B) Shown are results from GST-pull down assays using GST-fused to the C-termini of Shal1 (residues 411 to 571) and Shal2 (residues 411 to 490), then incubated with purified SKIP3 protein; glutathione-agarose beads alone (beads) and GST were used as negative controls. A representative immunoblot of the proteins pulled-down by GST-fusion proteins (top) shows that large amounts of SKIP3 are pulled-down by GST-Shal1C and GST-Shal2C, with much lower background levels pulled-down by beads alone or GST. Immunoblot was incubated with Ponceau S (PonsS) to confirm that GST protein(s) were indeed pulled-down by glutathione-beads. (C) SKIP encodes three alternatively spliced isoforms, SKIP1 (CG31163-PB/PC), SKIP2 (CG31163-PA), and SKIP3. Each isoform contains a different combination of protein binding domains, including two different SAM domains (orange) and an SH3 domain (green) domain. SKIP3 contains a unique eight residue C-terminal sequence (blue), which is required for binding Shal channels (see Fig. 2). (D) RT-PCR of SKIP3 from wild-type embryos. When RNA treated with DNase was used as template, SKIP3 was successfully reverse-transcribed and amplified by PCR (RT-PCR, left lane), suggesting that SKIP3 is a true isoform expressed in the embryo. When RNA was not treated with DNase, as indicated (−), SKIP3 could be amplified even without reverse transcription (middle lane), suggesting that DNase treatment is essential to degrade contaminating genomic DNA. A mock RT-PCR performed from DNase-treated RNA in the absence of reverse transcriptase (RT) confirmed that SKIP3 was not amplified without RT.