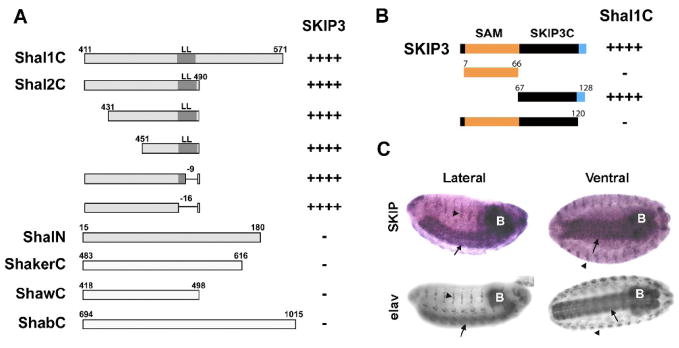
Fig. 2. Mapping the SKIP3-Shal Interaction, and Nervous System Specific Expression of SKIP3.
(A) Direct Y2H results are shown for “bait” fusion proteins, including C-terminal sequences from Shal1 (Shal1C), Shal2 (Shal2C), ShA1 (ShakerC), Shab-RB (ShabC), and Shaw2 (ShawC), and the N-terminus of Shal (ShalN), co-expressed with the SKIP3 “prey” fusion protein identified in our Y2H screen. Positive interactions, assayed by growth on stringent media and a blue color when grown on X-α-gal, are indicated by the number of “+” symbols; “−” indicates no/minimal growth or blue color. SKIP3 interacted specifically with the C-terminus of Shal. Binding is mediated by the sequence between residues 451 and 474 since deletions outside of this region, including the conserved di-leucine motif (LL), did not affect the interaction. (B) Direct Y2H results are shown for the Shal1C “bait” co-expressed with various truncations of SKIP3 as “prey”. Full-length SKIP3, containing a 60 amino acid N-terminal SAM domain (orange), followed by a 54 amino acid domain (SKIP3C; black), and a C-terminal eight amino acids (blue) unique to SKIP3, showed strong interaction with Shal1C, as expected. Protein-protein interaction, indicated by the number of “+” symbols, was assayed by growth on stringent media and a blue color when grown on X-α-Gal. Note that truncation of the C-terminal eight amino acids (blue) of SKIP3 completely disrupted interaction with Shal1C, while deletion of the SAM domain did not. Similar results were seen for Shal2C (data not shown). (C) Representative RNA in situ hybridization for SKIP (top) and immunostaining for the neuron-specific elav protein (bottom) in embryos 12 hours AEL. The single-stranded digioxygenin-labeled probe for in situs, amplified from SKIP3 template, likely hybridizes with both SKIP1 and SKIP3. After hybridization, the embryos were incubated with anti-Dig antibody and developed by NBT/BCIP. SKIP1/3 displays expression primarily in the central nervous system (brain indicated by “B”, ventral nerve cord indicated by arrows), and peripheral nervous system (indicated by arrowheads), similar to elav.