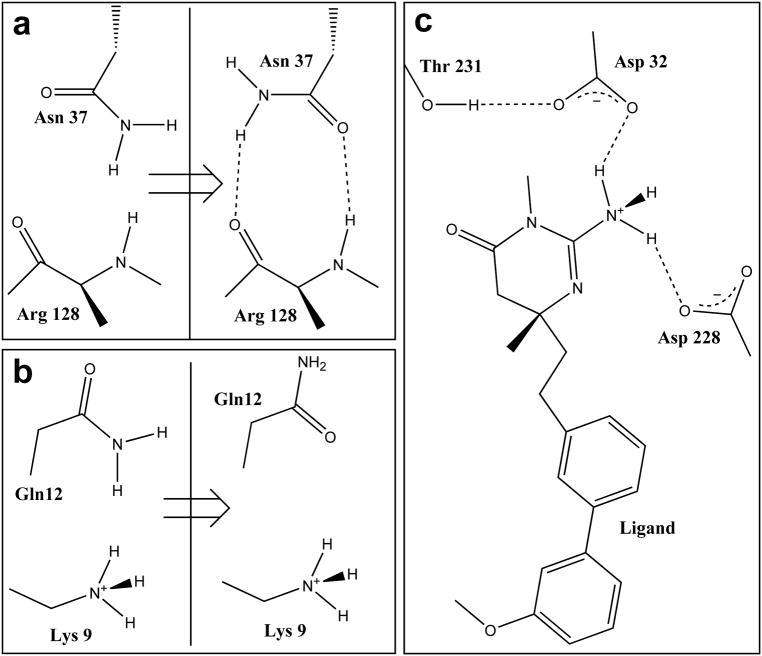
Figure 2.
Structural elements of the x-ray crystal structure of liganded β secretase (BACE-1, PDB code 2va7) as modified by computational titration. All residues with ionizable functional groups within 6 Å of the ligand were included in the titratable pool. Water molecules within 4 Å of atoms in both the ligand and protein were selected for optimization. a) The amide group of Asn37 is flipped 180° – improving its contact with backbone atoms of Arg128. b) Flipping the amide of Gln12 improves its interaction with Lys9. c) Protonating the ligand’s amine improves its interactions with both Asp32 and Asp228.