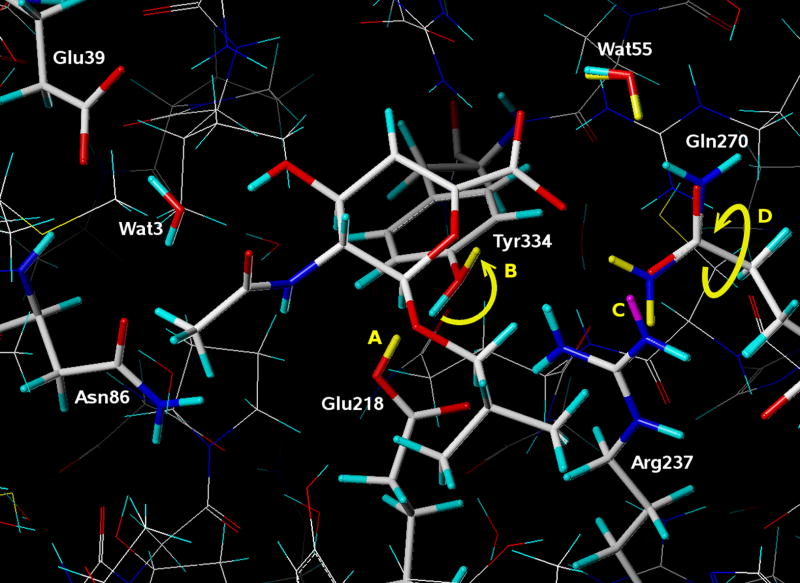
Figure 4.
A comparison between two models of the crystal structure of human sialidase NEU2 in complex with an isobutyl ether mimetic inhibitor (PDB 2f11). In one model hydrogen atoms for ionizable polar atoms were assigned conventionally (H atoms colored cyan and purple), using protonation states that are typically assumed for a neutral solution. In the other model both the protonation state for the ionizable polar groups and the resulting positions of ionizable hydrogens were optimized with computational titration. In both models the positions of hydrogens were energy minimized with molecular mechanics. Atoms colored yellow (added) and purple (deleted) indicate differences between the two models as labeled: A) Glu218 is protonated; B) the hydroxyl group on Tyr334 is rotated about 180°; C) Arg237 is deprotonated; and D) the amide group on the side chain of Gln270 is flipped.