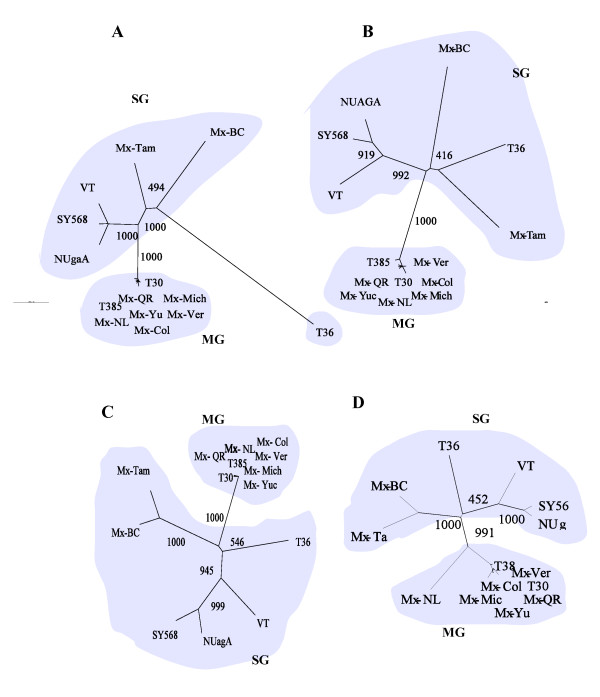
Figure 1.
Unrooted phylogenetic trees of the genomic regions p349-B/C (A), CP (B), p13 (C) and p23 (D) for 14 CTV isolates. The virus isolates that fell in separated groups are shaded and labeled with the group name (MG, mild group; SG, severe group). Trees were constructed by neighbor-joining method using nucleotide sequences aligned with Clustal × program and 1,000 bootstrap replications. Only the values over 400 are shown. The scale bar represents the number of nucleotide replacements per site: 0.01.